
ERα promotes SUMO1 transcription by binding with the ERE and enhances SUMO1-mediated protein SUMOylation in breast cancer
Highlight box
Key findings
• ERα-induced SUMO1 expression plays a key role in the regulation of BC proliferation.
What is known and what is new?
• E2 promoted the transcription and protein expression of SUMO1 via ERα binding to a 1/2ERE in the SUMO1 promoter.
• E2-ERα augmented SUMO1-mediated Ras SUMOylation and mediated cellular responses in ER-positive BC.
What is the implication, and what should change now?
• We need to pay attention to that SUMO1-induced protein SUMOylation, and elucidated its effect on cellular proliferation in BC.
Introduction
Breast cancer (BC) has one of the highest rates of cancer deaths among women worldwide, with long-term exposure to estrogen considered to be the principal factor leading to its tumorigenesis; estrogen-dependent BC accounts for approximately 2/3 of BC cases (1,2). In addition, clinical research using anti-estrogens or aromatase inhibitors to reduce local and distant recurrence has revealed that estrogen promotes the development of BC (3). The molecular mechanism underlying estrogen-induced BC is hypothesized to be mediated by the combination of estrogen and estrogen receptors (ERs) that bind to the estrogen-responsive element (ERE) of target-gene promoters or regulatory regions. ESR1 and ESR2 are different genes that encode ERα and ERβ subtypes, respectively (4), and elevated expression of ERα is observed in ER-positive BC and known to be related to BC growth. In addition, widespread expression of ERβ is found in BC, although its role remains unclear (5).
ERα is overexpressed in approximately 70% of BC cases (6). ERα is a ligand-activated transcription factor consisting of three functional domains of hormone binding, DNA binding and transcription activation. The ligand-binding domain (LBD) is recognized by the E2 (7). The deactivation domains AF-1 and AF-2 synergistically activate ERα. The DNA binding domain (DBD) recognizes estrogen response elements on DNA (8). ERα increases the expression level of carcinogenic proteins, including cyclin D1 and c-myc, and inhibits the level of cell cycle inhibitors, including P21 (9). ERα can also bind to promoters or regulatory regions of target genes that contain incomplete or truncated EREs and activate their transcription (10). E2-ERα signaling therefore plays a key role in the growth, migration, and invasion of BC cells (11). Because ERα and its signaling pathway play a crucial role in the development and progression of BC, anti-estrogen therapy and targeting ERα signaling pathway are important components of treatment for ERα positive BC patients.
SUMOylation is a vital post-translational modification that is critical to a variety of biologic functions, including cell growth, migration, and metastasis (12). SUMOylation is an enzyme cascade wherein small ubiquitin-related modifiers (SUMOs) are covalently bound to an internal lysine residue of a target protein by the carboxy-terminal glycine of processed SUMO (13). The binding of SUMO to proteins may thus be crucial to protein activity, subcellular localization, and stability (14). For example, increasing evidence reveals that SUMOylation can target various proteins, including nuclear transcription factors, membrane proteins, and cytoplasmic proteins, which are pivotal to BC progression (15). The dysregulation of SUMOylation could result in tumor progression, and is considered as a novel biomarker and possible therapeutic target for cancers (16). SUMO1, SUMO2, and SUMO3 are involved in the process of protein SUMOylation (17), and although investigators have previously identified a role for SUMO2 and SUMO3 in BC, the function for SUMO1 in BC remains arcane (18).
Accumulating evidence suggests that estrogen levels correlate with protein SUMOylation (19). It was reported that a tumor’s positive nuclear/negative cytoplasmic expression of SUMO proteins, including PIAS1, PIAS4, and UBC9, featured positive expression for ER, and that PIAS1, PIAS4, and UBC9 expressions were elevated in an ER-positive MCF-7 cell line compared with an ER-negative MDA-MB-436 cell line (20). Although E2-ER signaling is associated with protein SUMOylation in BC, the molecular mechanism that regulates SUMOylation remains largely undefined. In this study, we demonstrated that ERα activated SUMO1 gene transcription by binding with the ERE of the SUMO1 promoter in response to E2, and that ERα-induced SUMO1 expression was involved in BC development. We present this article in accordance with the ARRIVE reporting checklist (available at https://gs.amegroups.com/article/view/10.21037/gs-23-39/rc).
Methods
Cell culture
MDA-MB-231 and HeLa cells were cultured in DMEM (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS). T47D, MCF-7, and SK-BR-3 cells were incubated in RPMI-1640 (Gibco) containing 10% FBS. All cells were cultured at 37 ℃ in 5% CO2 in compressed air with high humidity.
Plasmids and transient transfection
The SUMO1-reporter construct and expression constructs have been described previously (21). Transient transfections were executed using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). After transfection for 24–48 h, luciferase activity was measured with a Luciferase Reporter Assay System (Promega).
Clinical samples
Tumor tissue samples were obtained from 86 ERα-positive BC patients who underwent surgery at the Affiliated Hospital of Nantong University. This study was approved by the institutional ethics committee of the Affiliated Hospital of Nantong University (ID: 2023-L084). Informed consent was obtained from each participant. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Protein extraction, immunoprecipitation, and western blot
Whole-cell protein extraction and immunoprecipitation were conducted as described previously (22), and western blot analysis used available antibodies. Primary antibodies were generated against SUMO1 (diluted 1:500, #ab227424, Abcam, Cambridge, UK), ERα (1/200, #ab32063, Abcam), Ras (1/5,000, #ab52939, Abcam), myc (1/1,000, #ab32072, Abcam), GST (1/1,000, #ab111947, Abcam), p-Akt (1/500, #ab8805, Abcam), Akt (1/500, #ab38449, Abcam), p-ERK1/2 (1/1,000, #ab278538, Abcam), ERK1/2 (1/10,000, #ab184699, Abcam), NF-κB p65 (1/1,000, #ab32536, Abcam), and phosphorylated (p-)NF-κB p65 (1/1,000, #ab86299, Abcam); GAPDH (1:10,000, #sc-47724, Santa Cruz Biotechnology, Dallas, TX, USA) was used as an internal control. The protein bands were visualized using an efficient chemiluminescence (ECL) detection kit (Thermo Fisher Scientific).
Real-time quantitative PCR (RT-qPCR)
RNA extraction was performed with TRIzol reagent (Invitrogen), and reverse transcription with a SuperScript First-Strand Synthesis System (Invitrogen). The RT-qPCR amplification was performed on an ABI Prism 7500 system. The relative RNA levels were calculated by the 2−ΔΔCt method.
Chromatin immunoprecipitation (ChIP) assay
We performed ChIP as described previously (23). Each immunoprecipitation reaction contained a small aliquot of lysate with 200 µg of protein, and PCR was used to amplify the precipitated genomic DNA.
Cell proliferation and colony formation assays
Cells were plated in 96-well plates at a density of 1×104 cells/well. We measured cellular proliferation by MTT assay according to the manufacturer’s protocol for a commercially available kit (KeyGen, Nanjing, China). For colony formation, cells were seeded at a density of 1,000 cells/well, and after 2 weeks, the colonies (>50 cells/colony) were fixed and stained with 0.1% crystal violet and photographed.
Orthotopic tumor model
Male nude mice (4–6 weeks old) were allocated to different groups, and 2×106 cells were injected into their right flanks. Four weeks later, the tumors were removed from the dead mice and photographed, and their volumes were calculated as length (mm) × width2 (mm2)/2. All nude mice are purchased from the Animal Center of Nantong University. This animal research protocol was approved by the Animal Ethics Committee of Nantong University (ID: P20230222-001), in compliance with institutional guidelines for the care and use of animals.
Statistical analysis
We used SPSS 20.0 to conduct statistical analyses. The differences between the control and experimental groups were analyzed using Student’s t-test and one-way analysis of variance (ANOVA). P<0.05 was deemed to be statistically significant.
Results
ERα induced SUMO1 expression in BC cells
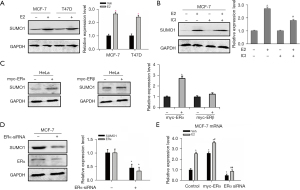
To evaluate the relationship between SUMO1 and ERs in BC, we examined whether E2 stimulated SUMO1 expression. The ER-positive MCF-7 and T47D cell lines treated with E2 exhibited increased SUMO1 protein levels in a dose- and time-dependent manner, but exerted no effect on SUMO2 or SUMO3 protein expression (Figure 1A, Figure S1A,S1B, Appendix 1). The same treatment of ER-negative MDA-MB-231 and SK-BR-3 cells had little effect on SUMO1 expression (Figure S1C). Treatment of MCF-7 and T47D cells with the anti-estrogen ICI 182780 partially reversed E2-enhanced SUMO1 expression (Figure 1B, Figure S1D). We also found that SUMO1 mRNA levels in MCF-7 and T47D cells were elevated in response to E2 in both dose- and time-dependent manner (Figure S1E). When we then assessed whether E2 could promote SUMO1 expression via ERα or ERβ using HeLa cells that did not contain measurable levels of ERα or ERβ but were transfected with ERα or ERβ, we found that overexpression of ERα increased SUMO1 expression, but that overexpression ERβ had no effect on HeLa cells (Figure 1C). Conversely, knockdown of ERα reduced the expression of SUMO1 in both MCF-7 and T47D cells (Figure 1D, Figure S1F). The effects of ERα on the regulation of SUMO1 transcription were thus further confirmed. As expected, we noted that knockdown of ERα downregulated SUMO1 transcription and that ERα overexpression increased SUMO1 transcription (Figure 1E, Figure S1G). Our results therefore proved that SUMO1 was a target gene of ERα transcription in BC.
ERα bound to SUMO1 promoter regions and regulated SUMO1 promoter activity
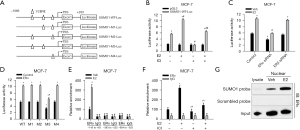
To further investigate the regulation of SUMO1 expression by ERα, the fragments containing the upstream and downstream regions of the transcriptional start site (TSS) were cloned and fused with the luciferase gene to produce a SUMO1-WT-Luc construct (Figure 2A). To examine whether E2 promoted the transcription of SUMO1, we determined the luciferase activities of MCF-7 and T47D cells transfected with SUMO1-WT-Luc after E2 or immune checkpoint inhibitor (ICI) treatment and observed that in the presence of E2 SUMO1-WT-Luc luciferase activity increased but that ICI lessened this effect (Figure 2B, Figure S2A). However, the level of luciferase activity expressed by SUMO1-WT-Luc was unaltered in response to E2 or ICI in MDA-MB-231 and SK-BR-3 cells (Figure S2A). When increasing amounts of ERα-expression plasmids were transfected into ERα-/ERβ-HeLa cells, SUMO1-WT-Luc showed a gene-dosage effect with ERα that was commensurate with the enhanced expression of luciferase activity (Figure S2B). However, the cells transfected with increasing levels of ERβ showed only a small increase in luciferase activity (Figure S2B). In addition, knockdown of ERα attenuated the reporter activity level with cellular exposure to E2 in MCF-7 and T47D cells, but exerted less of an effect in ERβ-knockdown cells (Figure 2C, Figure S2C). Taken together, these findings revealed that SUMO1 was regulated by the transcription of ERα. When we searched for ERα-responsive regions in the SUMO1 promoter, we located three 1/2EREs in the −564 to −553, −343 to −332, and −134 to −123 bp regions with respect to the TSS. To determine which portion of the promoter was most susceptible to ERα, we constructed truncated promoters (SUMO1-M1-Luc, SUMO1-M2-Luc, SUMO1-M3-Luc, and SUMO1-M4-Luc) and transfected them into MCF-7 cells to detect their effects on ERα-reactive activity. Luciferase activity was diminished when the −134 to −123 bp 1/2ERE was deleted, but was less affected when either the −564 to −553 or −343 to −332 bp region 1/2ERE was deleted; and the luciferase activity was increased with ERα overexpression (Figure 2D). Furthermore, when we transfected these luciferase vectors with ERα expression vectors into HeLa cells, we noted that overexpression of ERα elevated luciferase activity in MCF-7 and T47D cells, but that the increase in activity was less than in the −134 to −123 bp 1/2ERE-mutant transfected cells (Figure S2D). These findings indicated that this 1/2ERE was vital for ERα activity and drove the luciferase activity of SUMO1. We then implemented a ChIP assay to determine the binding of ERα to SUMO1 in MCF-7 cells, and demonstrated that when the cells were treated with E2, ERα only bound to region −145 to +63, but not to regions −664 to −523 or −383 to −252 (Figure 2E). We also noted binding of ERα to the SUMO1 promotor, and that E2 enhanced the binding between the SUMO1 probe and ERα, but reversed it with ICI pretreatment (Figure 2F). Besides, we observed that immunoprecipitated ERα bound to the nuclear extract incubated with a SUMO1 probe (Figure 2G). Taken together, our results demonstrated that endogenous ERα bound to the SUMO1 promoter and induced SUMO1 gene expression.
SUMO1 expression induced by ERα for BC cell growth
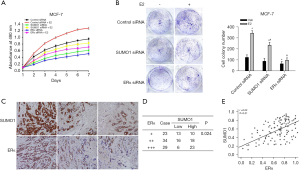
We assessed the effects of decreased SUMO1 expression on the biologic activities of BC cells. Treatment with E2 promoted the proliferation of MCF-7 and T47D cells with transfection of control siRNA, while in the presence of E2, inhibition of SUMO1 or ERα suppressed cellular proliferation (Figure 3A, Figure S3A). We then determined the effects of SUMO1 silencing on cell-colony formation, and showed that E2 treatment enhanced colony formation of MCF-7 and T47D cells, while in the absence or presence of E2, knockdown of SUMO1 decreased colony formation compared with the control siRNA-transfected group (Figure 3B, Figure S3B). Also, as we expected, downregulated expression of ERα reduced cell-colony formation, and these cells also lost their responsiveness to E2 (Figure 3B, Figure S3B). Our findings thus indicated that SUMO1 expression induced by ERα stimulated cellular proliferation. We also used an immunohistochemical (IHC) assay to ascertain the expression of and relationship between SUMO1 and ERα in ERα-positive BC samples (Figure 3C). With respect to ER-expression levels, 10 samples of SUMO1 expression were elevated in 23 ER+ patients, 18 samples were augmented in 34 ER++ patients, and 23 samples were high in 29 ER+++ patients (Figure 3D). When we analyzed the relationship between SUMO1 and ERα, we found that their expression levels correlated (Figure 3E), which suggested a correlation between SUMO1 and ERα in ERα-positive BC.
ERα induced SUMO1-mediated protein SUMOylation in BC cells
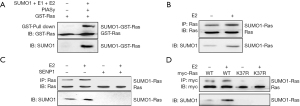
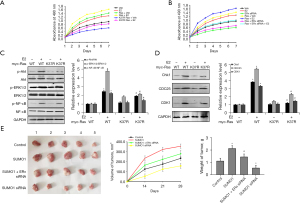
To further investigate the effects surrounding cell growth as mediated by SUMO1 expression and to clarify the underlying mechanism of action, we identified aspects of SUMO1-mediated protein SUMOylation. The SUMOylation mediated by SUMO1 was increased in the presence of E2 and reversed when SUMO1 and ERα were knocked down (Figure S4A), and vascular endothelial growth factor receptor (VEGFR) or HIF1α SUMOylation was also increased when cells were treated with E2 and reversed by ERα knockdown (Figure S4B, Figure S5). Immunoprecipitation combined with mass spectrometry was used to investigate novel target proteins of SUMO1, and we identified Ras as binding to SUMO1. Based on this interaction, we showed that Ras bound to SUMO1 both in vitro and in vivo, and that binding was enhanced in the presence of E2 and reduced with ERα knockdown (Figure 4A,4B). We then used SENP1 (24) (which removes SUMO from target proteins) and transfected it into cells, which revealed that SUMOylation of Ras was diminished when cells were pretreated with E2 (Figure 4C). We also identified the SUMOylation and binding site of Ras as Lys-37, and showed that mutation of this site to glycine decreased SUMOylation (Figure 4D). These findings indicated that in BC cells, ERα-induced expression of SUMO1 was involved in protein SUMOylation, and that Ras may indeed be the target in BC.
SUMO1-mediated protein SUMOylation associated with cellular proliferation in BC cells
Although investigators have determined that Ras occupies a critical role in cellular proliferation, whether its SUMOylation mediated by SUMO1 is involved in this process remains unknown. Because Ras overexpression in MCF7 cells increased their proliferation rate, we transfected mutant Ras into the cells to further ascertain the role of SUMOylation in the process, and observed that the proliferation induced by Ras was reversed when SUMOylation sites were mutated (Figure 5A). This finding indicated that Ras SUMOylation was essential to cell proliferation. When cells were treated with E2 or underwent ERα knockdown, Ras-mediated cell proliferation was enhanced in cells pretreated with E2, but was circumvented with ERα knockdown in the presence of E2 (Figure 5A,5B). We also evaluated signal transduction-pathway activation in these cells, and demonstrated that activation of RAS-mediated nuclear factor kappa B and CDK1 signaling was increased when cells were incubated with E2, and that it was further enhanced when the SUMOylation sites were mutated (Figure 5C,5D). To gain further insights into the ERα induction of SUMO1-mediated protein SUMOylation in BC, we examined tumor growth in BC in vivo. We found that the MCF-7 line that stably expressed SUMO1 grew more rapidly, that the cell line that stably expressed SUMO1 siRNA grew more sluggishly, and that SUMO1-induced tumor growth was suppressed with ERα knockdown (Figure 5E), findings that indicated that E2-ERα enhanced cellular proliferation via increased SUMO1-mediated RAS SUMOylation.
Discussion
Intriguingly, women who are chronically exposed to artificial light at night or are engaged in work that may change their circadian or menstrual rhythms, are more likely to develop BC, with E2 known to be a prominent factor in the effect (25), and numerous reports have shown that the tumorigenesis of BC is related to signaling pathways such as protein SUMOylation (26). It is therefore of paramount importance to analyze the signal transduction of estrogen, which is crucial to several cellular processes and to the pathogenesis of BC (27). In the present study, we found that E2-ERα activated SUMO1 expression at the transcriptional level, and that ERα-induced SUMO1 expression was involved in the proliferation of BC cells. In addition, SUMO1 can bind to Ras, inducing subsequent protein SUMOylation and BC development. Collectively, our findings indicated that SUMO1 was the target of ERα, and that protein SUMOylation regulated BC development (Figure S6).
ERα-positive BC can be controlled by ERα modulators such as tamoxifen (28). However, acquired resistance to tamoxifen is common, making it an important clinical issue in BC treatment. Understanding the dysregulation of ERα signals will help develop new strategies for treating cancer patients. A previous study has shown that the ubiquitin proteasome system (UPS) is involved in the regulation of ERα stability (29). E3 ubiquitin ligases induce 26S proteasome mediated ERα degradation by increasing polyubiquitin to ERα lysine residues (30). However, further research is needed to explore the exact mechanism of ERα dysfunction. We demonstrated that a higher proportion of ERα-positive BC samples possessed higher SUMO1 protein levels, indicating that SUMO1 transcription may be upregulated in ERα-positive tumors (31,32). Owing to the ambiguity in tumor grades recorded for the tissue samples, a correlation between tumor grade and SUMO1 expression could not be established. Timing of tumor resection may be key to studying the circadian rhythm that influences SUMO1 protein actions, but this endpoint was not fully recorded in our investigation (33). We concluded that the MCF-7 and T47D cell lines exhibited SUMO1 mRNA and protein levels that were specifically affected by ERα (34). The differences in E2 response between MDA-MB-231 (ERβ-positive/ERα-negative cells) and T47D cells (ERα-positive/ERβ-negative cells) revealed that ERβ may wield a less potent regulatory effect on the SUMO1 response to E2 than to ERα (35), and this was confirmed by the absence of changes in SUMO1 protein levels in T47D cells that overexpressed ERβ (36). These results were also consistent with the experimental data we collected using a reporter gene. ICI and E2 competed for ERα binding, promoting ERα degradation, and thus disrupted its location in the cell nucleus and subsequent dimerization.
Ras is a major oncogene in the mammalian Ras gene family, encoding a protein (RAS) that belongs to the small-GTPase superfamily, and plays an essential role in tumorigenesis (37-39). In this study, we found that Ras can complex with SUMO1 and promote cellular proliferation via enhanced SUMOylation, and we additionally identified the SUMOylation site of the two proteins. SUMOylation is essential in the development of BC, and several oncogenes have been identified in the SUMOylation process (40). Our findings have thus critically supplemented the available information regarding SUMO1-induced protein SUMOylation, and elucidated its effect on cellular proliferation in BC.
Conclusions
In conclusion, our findings reinforce the concept that ERα-induced SUMO1 expression is vital to the regulation of BC proliferation; and that targeting ERα-SUMO1 to attenuate protein SUMOylation may be a novel therapeutic inhibitor of BC development.
Acknowledgments
Funding: This work was supported by grants from
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://gs.amegroups.com/article/view/10.21037/gs-23-39/rc
Data Sharing Statement: Available at https://gs.amegroups.com/article/view/10.21037/gs-23-39/dss
Peer Review File: Available at https://gs.amegroups.com/article/view/10.21037/gs-23-39/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://gs.amegroups.com/article/view/10.21037/gs-23-39/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the institutional ethics committee of the Affiliated Hospital of Nantong University (ID: 2023-L084). Informed consent was obtained from each participant. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The animal research protocol was approved by the Animal Ethics Committee of Nantong University (ID: P20230222-001), in compliance with institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Harbeck N, Penault-Llorca F, Cortes J, et al. Breast cancer. Nat Rev Dis Primers 2019;5:66. [Crossref] [PubMed]
- Kalyanaraman A, Gnanasampanthapandian D, Shanmughan P, et al. Tamoxifen induces stem-like phenotypes and multidrug resistance by altering epigenetic regulators in ERα+ breast cancer cells. Stem Cell Investig 2020;7:20. [Crossref] [PubMed]
- Waks AG, Winer EP. Breast Cancer Treatment: A Review. JAMA 2019;321:288-300. [Crossref] [PubMed]
- Saha T, Makar S, Swetha R, et al. Estrogen signaling: An emanating therapeutic target for breast cancer treatment. Eur J Med Chem 2019;177:116-43. [Crossref] [PubMed]
- Zhou Y, Liu X. The role of estrogen receptor beta in breast cancer. Biomark Res 2020;8:39. [Crossref] [PubMed]
- Xu Y, Huangyang P, Wang Y, et al. ERα is an RNA-binding protein sustaining tumor cell survival and drug resistance. Cell 2021;184:5215-29.e17. [Crossref] [PubMed]
- Morrill GA, Kostellow AB, Gupta RK. Transmembrane helices in "classical" nuclear reproductive steroid receptors: a perspective. Nucl Recept Signal 2015;13:e003. [Crossref] [PubMed]
- Kotler E, Shani O, Goldfeld G, et al. A Systematic p53 Mutation Library Links Differential Functional Impact to Cancer Mutation Pattern and Evolutionary Conservation. Mol Cell 2018;71:178-90.e8. [Crossref] [PubMed]
- Zamir-Nasta T, Pazhouhi M, Ghanbari A, et al. Expression of cyclin D1, p21, and estrogen receptor alpha in aflatoxin G1-induced disturbance in testicular tissue of albino mice. Res Pharm Sci 2021;16:182-92. [Crossref] [PubMed]
- Kumar A, Dhillon A, Manjegowda MC, et al. Estrogen suppresses HOXB2 expression via ERα in breast cancer cells. Gene 2021;794:145746. [Crossref] [PubMed]
- Livezey M, Kim JE, Shapiro DJ. A New Role for Estrogen Receptor α in Cell Proliferation and Cancer: Activating the Anticipatory Unfolded Protein Response. Front Endocrinol (Lausanne) 2018;9:325. [Crossref] [PubMed]
- Han ZJ, Feng YH, Gu BH, et al. The post-translational modification, SUMOylation, and cancer Int J Oncol 2018;52:1081-94. (Review). [PubMed]
- Yang Y, He Y, Wang X, et al. Protein SUMOylation modification and its associations with disease. Open Biol 2017;7:170167. [Crossref] [PubMed]
- Seeler JS, Dejean A. SUMO and the robustness of cancer. Nat Rev Cancer 2017;17:184-97. [Crossref] [PubMed]
- Rabellino A, Khanna KK. The implication of the SUMOylation pathway in breast cancer pathogenesis and treatment. Crit Rev Biochem Mol Biol 2020;55:54-70. [Crossref] [PubMed]
- Lin Y, Wang Q, Lin Y, et al. An immunohistochemical panel of three small ubiquitin-like modifier genes predicts outcomes of patients with triple-negative breast cancer. Gland Surg 2021;10:1067-84. [Crossref] [PubMed]
- Kunz K, Piller T, Müller S. SUMO-specific proteases and isopeptidases of the SENP family at a glance. J Cell Sci 2018;131:jcs211904. [Crossref] [PubMed]
- Zhang Z, Liao B, Xu M, et al. Post-translational modification of POU domain transcription factor Oct-4 by SUMO-1. FASEB J 2007;21:3042-51. [Crossref] [PubMed]
- Knutson TP, Truong TH, Ma S, et al. Posttranslationally modified progesterone receptors direct ligand-specific expression of breast cancer stem cell-associated gene programs. J Hematol Oncol 2017;10:89. [Crossref] [PubMed]
- Alshareeda AT, Negm OH, Green AR, et al. SUMOylation proteins in breast cancer. Breast Cancer Res Treat 2014;144:519-30. [Crossref] [PubMed]
- Nanos-Webb A, Deyrieux A, Bian XL, et al. Cloning the human SUMO1 promoter. Mol Biol Rep 2010;37:1155-63. [Crossref] [PubMed]
- Li J, Wen S, Li B, et al. Phosphorylation-Mediated Molecular Pathway Changes in Human Pituitary Neuroendocrine Tumors Identified by Quantitative Phosphoproteomics. Cells 2021;10:2225. [Crossref] [PubMed]
- McColl H, Zagozewski JL, Eisenstat DD. Chromatin Immunoprecipitation (ChIP) Protocols for the Cancer and Developmental Biology Laboratory. Methods Mol Biol 2019;1869:143-53. [Crossref] [PubMed]
- Hotz PW, Müller S, Mendler L. SUMO-specific Isopeptidases Tuning Cardiac SUMOylation in Health and Disease. Front Mol Biosci 2021;8:786136. [Crossref] [PubMed]
- Figueroa JD, Gierach GL, Duggan MA, et al. Risk factors for breast cancer development by tumor characteristics among women with benign breast disease. Breast Cancer Res 2021;23:34. [Crossref] [PubMed]
- Rasha F, Sharma M, Pruitt K. Mechanisms of endocrine therapy resistance in breast cancer. Mol Cell Endocrinol 2021;532:111322. [Crossref] [PubMed]
- Dimitrakopoulos FI, Kottorou A, Tzezou A. Endocrine resistance and epigenetic reprogramming in estrogen receptor positive breast cancer. Cancer Lett 2021;517:55-65. [Crossref] [PubMed]
- Xu Y, Qin L, Sun T, et al. Twist1 promotes breast cancer invasion and metastasis by silencing Foxa1 expression. Oncogene 2017;36:1157-66. [Crossref] [PubMed]
- Tang J, Luo Y, Long G, et al. MINDY1 promotes breast cancer cell proliferation by stabilizing estrogen receptor α. Cell Death Dis 2021;12:937. Erratum in: Cell Death Dis 2022;13:88. [Crossref] [PubMed]
- Tang J, Wu Z, Tian Z, et al. OTUD7B stabilizes estrogen receptor α and promotes breast cancer cell proliferation. Cell Death Dis 2021;12:534. [Crossref] [PubMed]
- Payne AW, Pant DK, Pan TC, et al. Ceramide kinase promotes tumor cell survival and mammary tumor recurrence. Cancer Res 2014;74:6352-63. [Crossref] [PubMed]
- Nait Achour T, Sentis S, Teyssier C, et al. Transcriptional repression of estrogen receptor α signaling by SENP2 in breast cancer cells. Mol Endocrinol 2014;28:183-96. [Crossref] [PubMed]
- Ying S, Dünnebier T, Si J, et al. Estrogen receptor alpha and nuclear factor Y coordinately regulate the transcription of the SUMO-conjugating UBC9 gene in MCF-7 breast cancer cells. PLoS One 2013;8:e75695. [Crossref] [PubMed]
- Linher-Melville K, Singh G. The transcriptional responsiveness of LKB1 to STAT-mediated signaling is differentially modulated by prolactin in human breast cancer cells. BMC Cancer 2014;14:415. [Crossref] [PubMed]
- Patani N, Dunbier AK, Anderson H, et al. Differences in the transcriptional response to fulvestrant and estrogen deprivation in ER-positive breast cancer. Clin Cancer Res 2014;20:3962-73. [Crossref] [PubMed]
- Kirn V, Shi R, Heublein S, et al. Estrogen receptor promoter methylation predicts survival in low-grade ovarian carcinoma patients. J Cancer Res Clin Oncol 2014;140:1681-7. [Crossref] [PubMed]
- De Luca A, Maiello MR, D'Alessio A, et al. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin Ther Targets 2012;16:S17-27. [Crossref] [PubMed]
- Johnson DS, Chen YH. Ras family of small GTPases in immunity and inflammation. Curr Opin Pharmacol 2012;12:458-63. [Crossref] [PubMed]
- Kubota Y, O'Grady P, Saito H, et al. Oncogenic Ras abrogates MEK SUMOylation that suppresses the ERK pathway and cell transformation. Nat Cell Biol 2011;13:282-91. [Crossref] [PubMed]
- Karami S, Lin FM, Kumar S, et al. Novel SUMO-Protease SENP7S Regulates β-catenin Signaling and Mammary Epithelial Cell Transformation. Sci Rep 2017;7:46477. [Crossref] [PubMed]


