
Molecular profiles and circulating tumor-DNA detected in Chinese early stage breast cancer
Introduction
This article provides an overview of the incidence and mortality of cancer in the world and China in 2020 based on the most recent data compiled by the International Agency for Research on Cancer (IARC). The IARC reported that female breast cancer has now surpassed lung cancer as the most common cancer in the world and China (1,2). Thus, more attention needs to be paid to its occurrence and development.
Breast cancer is a highly heterogeneous disease with diverse tissue morphology and molecular subtypes, each of which has different molecular characteristics, prognoses, and responses to therapies (3,4). Based on the expression of Ki67, 5 molecular types of breast cancer have been identified, and more subtypes remain to be reported (5,6). According to immunohistochemical or fluorescence in situ hybridization markers, breast cancer consists of the following 3 major tumor subtypes: estrogen receptor (ER) or progesterone receptor (PR) expression and Erb-B2 receptor tyrosine kinase 2 [ERBB2; human epidermal growth factor receptor 2 (HER2)] gene expression or amplification. Many studies have shown the that 3 subtypes have different prognoses and treatment strategies (7,8). However, with the development of gene-sequencing technology, more and more genome biomarkers [e.g., BReast CAncer gene 1 (BRCA1), BReast CAncer gene 2 (BRCA2), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), and cyclin-dependent kinase 4/6 (CDK4/6)], and immunomarkers [e.g., tumor-infiltrating lymphocytes, and programmed death-ligand 1 (PD-L1)] have been reported (8).
8 targeted therapy drugs for metastatic breast cancer have been approved, including trastuzumab for HER2+ patients, and Olaparib for germline BRCA mutation patients (7). However, the treatments of early stage and locally advanced breast cancer mainly include surgical resection, neoadjuvant chemotherapy or adjuvant radiotherapy or chemotherapy (9). Unlike advanced breast cancer, early stage breast cancer focuses more on the risk assessment of postoperative recurrence and metastasis. Exploring the key mutation events in the occurrence and development of early breast cancer may be conducive to the discovery of biomarkers, which are beneficial to the early screening and prevention of breast cancer.
In relation to the genomic mutations, tumor protein (TP53) and PIK3CA have been implicated in the development of breast cancer (10). Some studies have reported differences in the genomic mutation profiles of breast cancer between Chinese and Western patients. Young Chinese breast cancer patients (aged ≤35 years old) have higher mutation rates of TP53 (51% vs. 30%) and ERBB2 (34% vs. 24%) than young Western breast cancer patients; however, the PIK3CA (25% vs. 30%) and GATA3 (15% vs. 24%) mutation frequency is significantly lower in Chinese breast cancer patients than Western breast cancer patients (11).
Circulating tumor DNAs (ctDNAs) are some free DNA fragments produced by tumor cells in the process of necrosis or apoptosis. CtDNA carries the complete information of gene mutations in tumors or metastatic lesions, and these mutations will undergo some changes during treatment. Therefore, dynamic monitoring of ctDNA mutations can indicate treatment response to a certain extent. Based on high-depth and high-coverage of targeted next-generation sequencing (NGS), it can detect and analyze low-frequency variants of ctDNA. Therefore, the application of ctDNA to early detection and diagnosis of cancer is of great value. In breast cancer, recent studies have shown that ctDNA plays an important role in identifying micrometastasis and predicting the risk of recurrence, evaluating resistance to treatment or revealing tumor heterogeneity (12,13).
To better understand the genomic differences in breast cancer between Chinese and Western patients, we retrospectively analyzed the genomic data of tumor tissues and blood samples of 20 Chinese early invasive ductal breast cancer patients. Combined with the patients’ clinical information, a comprehensive analysis was carried out based on tissue mutation maps, tumor signaling pathways, the factors affecting lymph node metastasis, and the detection of circulating tumor deoxyribonucleic acid (ctDNA) in postoperative blood. The results of our analyses expected to provide insights into early stage breast cancer. We present the following article in accordance with the MDAR reporting checklist (available at https://gs.amegroups.com/article/view/10.21037/gs-21-691/rc).
Methods
Study design and patients
The data of 20 breast cancer patients who underwent next generation sequencing (NGS) at Geneplus-Beijing (Beijing, China) were analyzed. The study was approved by the Ethics Committee of The First Affiliated Hospital of Soochow University (No. 2022013) , and each patient provided their informed consent. All procedures were conducted in accordance with the Declaration of Helsinki (as revised in 2013).
DNA sequencing
Paired pretreatment tissue samples and plasma samples were used to identify mutations with a NGS panel containing 1,021 cancer-associated genes. As previously reported (14), we use TruSeq DNA kits to prepare genomic DNA (gDNA) sequencing libraries. Subsequently, DNA sequencing was performed on the CN 500 sequencing instrument (Illumina, San Diego, CA, USA). The reads were aligned to the human genome build Genome Reference Consortium (GRC) h37 using a Burrows-Wheeler aligner (15). MuTect2 (3.4-46-gbc02625) was used to identify single nucleotide variants (SNVs) (16), while Genome Analysis Toolkit (GATK) was employed to identify small insertions and deletions (indels) (17). All final candidate variants were verified using an integrative genomics viewer browser. Tumor mutational burden (TMB) was calculated as the number of somatic non-synonymous SNVs (indels per Mb in the coding region, with a variant allele fraction of ≥0.03).
Postoperative blood samples were collected 3–7 days after surgery. The blood samples were collected in ethylenediaminetetraacetic acid vacutainer tubes (BD Diagnostics, NJ, USA) and were subjected to the laboratory process within 3 h. DNeasy 19 and QIAamp (Qiagen) kits were used to extract gDNA from leukocytes and circulating free DNA (cfDNA) from plasma, and then KAPA DNA kits (Kapa Biosystems, MA, USA) were used to construct sequencing libraries. Next, DNA sequencing was performed as described above.
Statistical analyses
Differences in patients’ demographics were evaluated using the Fisher t-test. The other statistical analyses were performed using GraphPad Prism 8.0.2 (GraphPad Software, Inc.), and differences were evaluated using a 2-tailed unpaired Mann-Whitney U test. P values <0.05 were considered statistically significant.
Results
Patient cohort
Twenty patients, who had been diagnosed with breast cancer, were enrolled in this study. The characteristics of the patients are summarized in Table 1. The age of the patients ranged from 33 to 79 years (median: 42 years). 50% of patients had the hormone receptor positive (HR+) subtype, 4 patients had the triple-negative subtype, and 6 patients had the HER2 positive subtype. Tumor size ranged from 1.3 to 5 cm. Patients were distributed across stages I/II/III and most patients had lymph node metastasis. 3 patients had a family history of breast cancer, but none of the patients had a family history associated with hereditary breast cancer ovarian cancer syndrome. All lesions were invasive ductal carcinomas. None of the patients had received any treatment other than surgery.
Table 1
| Characteristics | N=20 |
|---|---|
| Age, median [range] years | 42 [33–79] |
| Subtype | |
| HR+/HER2− | 10 (50%) |
| TNBC | 4 (20%) |
| HER2+ | 6 (30%) |
| Tumor size, median [range], cm | 2.25 [1.3–5] |
| Stage | |
| I | 6 (30%) |
| II | 8 (40%) |
| III | 6 (30%) |
| N stage | |
| N0 | 9 (45%) |
| N1/N2 | 11 (55%) |
| Family history | 3 (15%) |
| Histological type | |
| Invasive ductal carcinoma | 20 (100%) |
HR+, hormone receptor positive; HER2 (also known as ERBB2), human epidermal growth factor receptor 2; TNBC, triple negative breast cancer.
Molecular characteristics of patients
Primary tumor samples from the 20 patients were sequenced to identify somatic mutations. Mutations were identified in all patients. Seventeen patients (85%) had multiple mutations, and only 3 patients (15%) had 1 mutation. The median allele frequency was 15.7%; 142 somatic mutations were identified, including 77 somatic base substitutions, 12 small indels, 49 copy number variations and 4 rearrangements. As Figure 1A shows, the most frequently altered genes were TP53 (70%), PIK3CA (40%), MYC (35%), ERBB2 (30%), and CDK12 (20%).
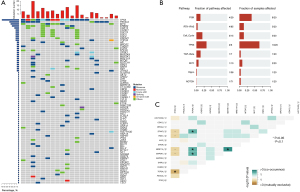
The consistency of ERBB2 amplification with NGS with the results of clinical immunohistochemical detection of HER2 was as high as 100%. Six (30%) patients were detected to have both ERBB2 amplification and to be HER2 positive, and these two markers were simultaneously detected were common in the breast cancer patients without lymph node metastasis (5/6, 83.3%) and in stages IA–IIA. In addition, HER2+ patients had a higher incidence of concomitant mutations than the HER2− patients, such as MYC, Cyclin D1 (CCND1), or fibroblast growth factor (FGF) family gene amplification, PIK3CA or ERBB2 SNV. TMB-H was detected in 1 of the HER2+ patients, and another patient a novel gene rearrangement of RARA-MYO15B. Additionally, 1 patient with the triple-negative breast cancer (TNBC) subtype had 3 novel gene rearrangements, comprising ETV6-PRB2 [intergenic, E26 transformation-specific variant 6 gene (ETV6), The retinoblastoma protein pRb and its relatives pRb2 (p130)], NOTCH1-PMPCA [intergenic, Notch is an ancient signaling pathway that includes Notch 1, mitochondrial processing peptidase-αprotein (PMPCA)], and EGFR-LRP6. Eight (40%) patients had the PIK3CA mutation (5 had the HR+, 2 had HER2+, and 1 had the TNBC subtype). Six patients had PIK3CA hotspot mutations in the helical domain (p.H1047R), and the other 2 mutations occurred in other domains (p.E418_L422delinsV and p.E545A).
A pathway analysis was conducted of all the mutant genes. Most of the mutated genes clustered in the TP53 pathway, followed by the PI3K pathway. Among the genes involved in the PI3K pathway, the most commonly mutated were PIK3CA (62%) and Mammalian target of rapamycin (MTOR) (15%). Among the genes involved in the RTK/RAS (Receptor Tyrosine Kinase/Rat sarcoma) pathway, the most commonly mutated were Mitogen-Activated Protein Kinase Kinase 4 (MAP2K4) (50%) and Mitogen-Activated Protein Kinase Kinase Kinase 1 (MAP3K1) (50%). Among the cell cycle pathway-related genes, the most commonly mutated were MYC (39%), CDK12 (22%), and CCND1 (17%; see Figure 1B). The co-existence of mutations detected in 20 patients was also analyzed. TOP2A was found to be mutually exclusive with TP53 (P<0.05), and to co-occur with MAP3K1, Zinc Finger Homeobox 3 (ZFHX3), and MRE11 Homolog A (MRE11A) (P<0.05) (Figure 1C).
Next, the TMB was analyzed. A previous research study reported that the median TMB was 2.63 mut/Mb in primary or metastatic breast cancer (18). Another study reported that the mean TMB was 6.28 mut/Mb in advanced or metastatic breast cancer (19). However, in our cohort, the TMB was lower than metastatic breast cancer, and ranged from 0 to 9.6 mut/Mb (median: 1.92 mut/Mb). The TMB of the TNBC subtype was numerically, but not significantly, higher than the TMBs of the HR+ (4.8 vs. 0.96; P=0.315) and HER2+ (4.8 vs. 1.92; P=0.769) subtypes. Our results are consistent with those reported in another study (18).
Subsequently, the genomic characteristics were analyzed. Germline mutations were found in 3 (0.15%) patients, including 2 BRCA2 mutations (1 pathogenic and 1 likely pathogenic) and 1 RAD51D pathogenic mutation. All 3 patients were young (aged 33, 45, and 47 years old) and 2 patients had TNBC subtypes. Additionally, 1 patient had a family history (that patient’s father had a stomach or gastroesophageal junction tumor). Of these 3 patients, ctDNA was detected in 1 patient’s postoperative peripheral blood sample.
Correlation between driver mutation and clinical characteristics
The therapeutic targeting of abnormal cancer driver mutations is garnering increased attention. Tumors with driver mutations were profiled in terms of their clinical and genomic characteristics. PIK3CA is a common driver mutation in breast cancer, especially in the HR+ subtype. As stated above, 8 of the 20 (40%) patients in this study presented with the PIK3CA mutation. The patients were divided into a PIK3CA mutation group and a wildtype group. The PIK3CA mutation group had a higher age (47 vs. 40; P=0.14) and a higher proportion of the patients in the PIK3CA mutation group were in stage I/II (85.7% vs. 69.2%; P=0.24) compared to the wildtype group; however, the differences were not significant. The PIK3CA wildtype group had a numerically higher proportion of patients with lymph node metastasis (61.5% vs. 42.9%; P=0.62) and a higher TMB (2.5 vs. 2; P=0.28) than the mutation group; however, the difference was not significant.
The relationship between the ERBB2 mutation and clinical and genomic characteristics were also analyzed. The ERBB2 mutation group had a numerically higher age (60 vs. 40; P=0.15), a higher TMB (2.4 vs. 1.4; P=0.51), and a higher proportion of patients in I/II stage (100% vs. 42.9%; P=0.26) compared to the wildtype group. Notably, the ERBB2 wildtype group had a significantly higher proportion of patients with lymph node metastasis than the ERBB2 mutation group (71.4% vs. 16.7%; P=0.049).
The essential role of BRCA1 and BRCA2 proteins in homologous recombination repair (HRR), which is a high-fidelity DNA double-strand break (DSB) repair mechanism, has been extensively documented. In this study, we also analyzed the HRR mutation. HRR mutations were found in 12 patients (5 had the HR+ subtype, 2 had the TNBC subtype, and 5 had the HER2+ subtype). No association was found between HRR mutations and clinical or genomic characteristics.
Genomic differences between patients with and without lymph node metastasis were then compared. Figure 2 sets out the top 10 frequently altered genes in the 2 groups. The TP53 and PIK3CA genes were highly altered in both groups. Further, both ERBB2 mutations (55.6% vs. 9.1%; P=0.049) and high-frequency CDK12 mutations (44.4% vs. 0%; P=0.026) were found in patients without lymph node metastasis, significantly difference to patients with lymph node metastasis.
Correlation between ctDNA and patient characteristics
Postoperative blood was obtained from all patients, and plasma DNA was extracted to analyze the presence of ctDNA. Buffy coat DNA was analyzed to control for the clonal hematopoiesis of indeterminate potential. ctDNA was detected in 7 of the 20 patients (35%), at a median allele frequency of 0.63%. Previous studies have shown that the detection of ctDNA in postoperative blood is associated with a higher risk of recurrence for early breast cancer (20,21). Thus, we investigated whether ctDNA detection was associated with any characteristics at diagnosis.
As Table 2 shows, age and subtype were not associated with ctDNA detection. Next, tumor size was compared in both the positive and negative groups. Interestingly, the majority of patients in whom ctDNA was detected were in stage I/II (71.4%). Similarly, driver mutations were not associated with the detection of ctDNA; 85.7% and 71.4% patients detected to have ctDNA had PIK3CA or ERBB2 wildtype, respectively. HRR mutations may be associated with ctDNA detection, but the association was not significant (P=0.085). Subsequently, we observed that patients positive for ctDNA had a higher TMB when grouped by the median TMB (1.92 mut/Mb) than those negative for ctDNA. We also found that patients positive for ctDNA had a higher TMB than patients negative for ctDNA (85.7% vs. 38.5%; P=0.043). These results demonstrated that the detection of ctDNA in postoperative blood is not associated with clinical characteristics but is associated with the TMB of patients.
Table 2
| Characteristics | Positive (n=7) | Negative (n=13) | P value |
|---|---|---|---|
| Age, years | |||
| Median | 43 | 40 | 0.7123 |
| Mean ± SD | 47.5±4.83 | 46±3.71 | |
| >42 year, n (%) | 4 (57.1) | 6 (46.2) | 1 |
| Subtypes, n (%) | 0.376 | ||
| Luminal A | 0 (0) | 3 (23.1) | |
| Luminal B | 2 (28.6) | 5 (38.4) | |
| HER2+ | 3 (42.8) | 3 (23.1) | |
| TNBC | 2 (28.6) | 2 (15.4) | |
| Tumor size | |||
| Median | 2.3 | 2.2 | 0.876 |
| Mean ± SD | 2.54±0.46 | 2.72±0.32 | |
| >2.25 cm, n (%) | 4 (57.1) | 6 (46.2) | 1 |
| Stage, n (%) | 0.612 | ||
| I/II | 5 (71.4) | 9 (69.2) | |
| III | 2 (28.6) | 4 (30.8) | |
| N-stage, n (%) | 1 | ||
| N0 | 3 (42.9) | 6 (46.2) | |
| N1/N2 | 4 (57.1) | 7 (53.8) | |
| PIK3CA, n (%) | 0.328 | ||
| mut | 1 (14.3) | 6 (46.2) | |
| wt | 6 (85.7) | 7 (53.8) | |
| ERBB2, n (%) | 1 | ||
| mut | 2 (28.6) | 3 (23.1) | |
| wt | 5 (71.4) | 10 (76.9) | |
| HRR, n (%) | 0.085 | ||
| mut | 6 (85.7) | 6 (46.2) | |
| wt | 1 (14.3) | 7 (53.8) | |
| TMB | |||
| Median | 2.88 | 0.97 | 0.17 |
| Mean ± SD | 3.84±1.19 | 2.58±0.81 | |
| >1.92 mut/Mb, n (%) | 6 (85.7) | 5 (38.5) | 0.0427 |
HER2 (also known as ERBB2), human epidermal growth factor receptor 2; TNBC, triple negative breast cancer; mut, mutation; wt, wild type; HRR, homologous recombination repair; TMB, tumor mutational burden.
Discussion
Breast cancer is the most frequently diagnosed cancer worldwide (22). Approximately 95% of women presenting with early stage breast cancer do not have macroscopic metastatic disease (22). In the preceding decades, molecular characterization has revolutionized breast cancer research and therapeutic approaches. In this study we examined the genomic characteristic of Chinese early stage breast cancer Patients. Additionally, the relationship between ctDNA detection in postoperative blood and patients’ characteristics was also described.
As Xiao et al. reported, TP53 (47.0%), PIK3CA (45.0%), and ERBB2 (30.0%) are the most frequently mutated genes, but other gene variations, such as CDK12 (18.0%), also occur (11). In our study, the most frequently altered genes were TP53 (70%), PIK3CA (40%), MYC (35%), ERBB2 (30%), and CDK12 (20%). Overall, the detection rate of high-frequency mutation genes in early breast cancer was relatively consistent. Xiao et al. also found that ERBB2 aberrations were more likely to co-occur with CDK12 [odds ratio (OR) =10]. Similarly, 25% (5/20) of patients were found have both the ERBB2 and CDK12 aberrations in our study.
Breast cancer is not a single entity; rather, it comprises multiple subtypes, each with its own set of genomic characteristics and signatures (23). The PI3K pathway is commonly altered in breast cancer and contributes to the development of breast cancer (24). In our cohort, 40% of patients had PI3K pathway mutations. Consistent with previous findings (22), TP53 was the most common mutation in this study, followed by PIK3CA. The PIK3CA gene encodes for the α-isoform of the catalytic subunit (p110α) of class IA PI3K kinase. PIK3CA somatic mutations have been reported in around 20–40% of early breast cancer patients (25,26). In this study, 40% of the patients had PIK3CA mutations. Additionally, similar to previous studies (27), we found that the hotspot of PIK3CA was p.H1047R. As André et al. (28) reported, switching from PIK3CA-wildtype to PIK3CA mutations was very rare, which could explain the stable incidence between early and late stage breast cancer. Further, another study showed that PIK3CA mutations were associated with older age (29). However, while we found that patients PIK3CA mutations were somewhat older, they were not significantly older than the other patients.
Genomic features were compared between patients with and without lymph node metastasis. Interestingly, we found a higher proportion of ERBB2 and CDK12 mutations in patients without lymph node metastasis. However, a study of 589 Chinese patients with early stage breast cancer showed ERBB2 and CDK12 gene amplification were more enrichment in the lymph node metastasis group (11). Our study has some limitations. The sample size limited our analysis. Further, this single-center study, which was conducted in the Suzhou province, only included patients from southern China, which may have resulted in patient selection bias. Further research needs to be conducted to verify our findings. Specifically, a multi-center study should be conducted throughout China with a larger cohort.
In addition to TNBC, HR+ and HER2+ breast cancers are considered immunologically “cold” due to their low tumor-infiltrating lymphocyte infiltration (30). Additionally, the efficacy of single-agent therapy with pembrolizumab was evaluated in ER+/HER2− advanced breast cancer. Only 20% of ER+ breast cancer cells express the PD-L1 immune-checkpoint protein, and single-agent immune-checkpoint inhibitors (ICIs) have shown limited efficacy (objective response rate: 12%, 3/25) in treating ER+ tumors (31). Previous studies have shown that TNBC has a higher PD-L1 expression; however, only about 10% of breast cancers show PD-L1 expression. Compared to other tumor types, PD-L1 expression is not a valid predictive biomarker of ICI efficacy in breast cancer.
In recent years, the TMB has become a prominent independent efficacy prediction biomarker of ICIs in several cancer types, including lung cancer, melanoma, and endometrial cancer; however, the significance of the TMB in breast cancer remains unclear (32). Barroso-Sousa et al. reported that high TMB (18%) was associated with significantly longer PFS (12.5 vs. 3.7 months; P=0.04) in patients with metastatic TNBC (mTNBC) which received anti-PD-1/L1 inhibitors alone or combined with targeted therapy or chemotherapy (33). In another study, an immune-related prognostic score was established in 22 breast cancer cohorts with a total of 6,415 samples and developed a prognostic scoring system indicative of immune infiltration. Fifty-four immune cell types with the P<0.05 were identified as prognostic factors. Of these, 51 immune cell types with the HR <1 indicated that the higher levels of normalized enrichment score (NES) values were associated with longer survival time (34). One study enrolled 5,112 patients for RNA analysis to quantify 22 tumor-infiltrating immune cells (TIICs) in primary BC. The immune cell infiltration-based immune score model can be effectively and efficiently used to predict the prognosis of BC patients as well as the effect of chemotherapy (35). A study demonstrated that the median TMB was 2.63 mut/Mb; however, the TMB varied significantly according to the tumor subtype (HR−/HER2− > HER2+ >HR+/HER2−; P<0.05) and sample type (metastatic > primary; P=2.2×10-16) in primary (2,455/3,969, 61.9%) or metastatic (1,496/3,969, 37.7%) breast cancer (18). In advanced or metastatic breast cancer, the mean TMB was 6.28 mut/Mb (19). However, for early stage breast cancer, a study of Chinese breast cancer patients revealed that the median value of the TMB level was 4 mutations/Mb, and only 5.7% of patients had a TMB ≥10 mutations/Mb (11). In our study, a lower TMB level was observed; the median TMB was only 1.92 mut/Mb. However, the number samples in our study was small. Additionally, differences in the targeted-sequencing panel and the calculation method used to determine the TMB may have led to biases and inconsistencies in the data results. In all subtypes, the TNBC subtype had a numerically higher TMB than other subtypes; however, the differences were not significant. The correlation between driver mutations and the TMB was also analyzed. However, as previously reported (36), no associations were found between the TMB and PIK3CA, ERBB2, and HRR mutations.
Recently, ctDNA has been suggested to be a tumor biomarker because of its sensitivity, stability and specificity. However, the best method to measure ctDNA is uncertain. Digital PCR (dPCR) can only measure one or few known mutations. But NGS can measure many mutations simultaneously and can identify resistant clones newly emerged during treatment (13). An early lung cancer ctDNA detection study showed that, excluding technical limitations, there are mainly two factors that affect the consistency of ctDNA and tumor tissue detection. On the one hand, biological factors, such as the blood state of ctDNA before surgery, are easier to detect for Shedder. The pathological type such as lung squamous cell carcinoma is easier than lung adenocarcinoma. The higher Ki67, the easier to release DNA. Patients with lymphatic vascular invasion of the tumor are more likely to detect ctDNA. On the other hand, it is related to clinical treatment, such as detection time after surgery and after radiotherapy or chemotherapy. DYNAMIC studies have shown that the detection of ctDNA at 3 days after surgery is more likely to indicate recurrence than at 1 day after surgery (37). One of our study used the 1,021 panel have confirmed that blood ctDNA is highly consistent with somatic mutations in tumor tissues in patients with early breast cancer, and demonstrated that ctDNA can be used to predict tumor response to neoadjuvant chemotherapy (NAC) and prognosis (13). In addition, ctDNA can be used to predict and monitor therapeutic response in advanced breast cancer. Reported studies have confirmed that molecular tumor burden index (mTBI) indicators based on ctDNA detection can be used as predictors of disease progression. The results show that mTBI can evaluate the therapeutic efficacy and clinical imaging of 11 patients with metastatic breast cancer, and the mTBI of 6 patients was 8–16 weeks earlier than imaging, suggesting clinical disease progression (38). Another study has demonstrated that plasma PIK3CA ctDNA mutation is associated with clinical outcome in advanced breast cancer. The ctDNA analysis found PIK3CA p.H1047R mutation was more frequent in HER2+ disease and associated with worse OS. It was also the only mutation associated with shorter PFS through a multivariate analysis of HER2+ patients who were treated with trastuzumab (39). In the ctDNA study for breast cancer, researchers evaluated the possibility of using plasma ctDNA to monitor minimal residual disease (MRD) in breast cancer. The studies have found that continuous monitoring of gene mutations in plasma improves the sensitivity of predicting recurrence, and can predict recurrence 7.9 months earlier than clinical testing. Compared with primary tumor tissue sequencing, deep sequencing of plasma ctDNA can more accurately predict metastasis and recurrence (40). Takeshita et al. analyzed 253 plasma samples from 119 HR-positive breast cancer patients, and detected cfDNA before, after, and during follow-up. It was found that patients with an increased frequency of ESR1 gene mutations in cfDNA before and after treatment progressed in a shorter treatment time. ESR1 gene mutation is an effective predictor of treatment effect for hormone-positive metastatic breast cancer patients (41). As a biomarker, we found an association between the detection of ctDNA and a higher risk of recurrence for early breast cancer patients (20,21). We also found that the presence of ctDNA was not associated with the characteristics of patients. In particular, ctDNA was not more likely to be detected in patients with stage III than patients with I/II (10% vs. 35%). As previously reported (42), pre-operative ctDNA positivity of gastric cancer was associated with disease stage, and the number of patients that were ctDNA positive in stage III was greater than that in stages I and II (68% vs. 21%; P=0.0044). Patients with a higher tumor stage or lymph node involvement were more likely to have detectable ctDNA (P=0.005 and P=0.029, respectively). In addition, tumor volume was associated with ctDNA positivity (median 9 cm3 in the ctDNA positive group vs. 4.5 cm3 in the ctDNA negative group; P=0.0582). In our study, the tumor volume of 1 stage IIA patient, whose postoperative ctDNA was positive, was 12.5 cm3. Few studies have examined the relationship between postoperative ctDNA and early stage breast cancer. However, the TMB was found to be related to ctDNA detection. This study had a few limitations. The association between survival and ctDNA needs to be further analyzed. We plan to reanalyze our data when the postoperative follow-up data are more mature. Further research needs to be conducted to verify our findings with a larger cohort.
In conclusion, our study described that genomic characteristics of early stage breast cancer in Chinese patients. We found that the TMB was related to the detection of ctDNA in postoperative blood. Our findings provide novel insights into the characteristics related to genetic mutations and postoperative ctDNA.
Acknowledgments
Funding: The present study was supported by the Project of Nature Science Foundation of China (No. 81702048).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://gs.amegroups.com/article/view/10.21037/gs-21-691/rc
Data Sharing Statement: Available at https://gs.amegroups.com/article/view/10.21037/gs-21-691/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://gs.amegroups.com/article/view/10.21037/gs-21-691/coif). MXZ is an employee of Geneplus-Beijing. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures were conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of The First Affiliated Hospital of Soochow University (No. 2022013) . Written informed consent was provided by all the participants in the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Cao W, Chen HD, Yu YW, et al. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl) 2021;134:783-91. [Crossref] [PubMed]
- Nguyen QH, Pervolarakis N, Blake K, et al. Profiling human breast epithelial cells using single cell RNA sequencing identifies cell diversity. Nat Commun 2018;9:2028. [Crossref] [PubMed]
- Dong Y, Van Tine BA, Oyama T, et al. Taspase1 cleaves MLL1 to activate cyclin E for HER2/neu breast tumorigenesis. Cell Res 2014;24:1354-66. [Crossref] [PubMed]
- Eroles P, Bosch A, Pérez-Fidalgo JA, et al. Molecular biology in breast cancer: intrinsic subtypes and signaling pathways. Cancer Treat Rev 2012;38:698-707. [Crossref] [PubMed]
- Ali HR, Rueda OM, Chin SF, et al. Genome-driven integrated classification of breast cancer validated in over 7,500 samples. Genome Biol 2014;15:431. [Crossref] [PubMed]
- Waks AG, Winer EP. Breast Cancer Treatment: A Review. JAMA 2019;321:288-300. [Crossref] [PubMed]
- Loibl S, Poortmans P, Morrow M, et al. Breast cancer. Lancet 2021;397:1750-69. [Crossref] [PubMed]
- Thomas M, Kelly ED, Abraham J, et al. Invasive lobular breast cancer: A review of pathogenesis, diagnosis, management, and future directions of early stage disease. Semin Oncol 2019;46:121-32. [Crossref] [PubMed]
- Lang GT, Jiang YZ, Shi JX, et al. Characterization of the genomic landscape and actionable mutations in Chinese breast cancers by clinical sequencing. Nat Commun 2020;11:5679. [Crossref] [PubMed]
- Xiao W, Zhang G, Chen B, et al. Characterization of Frequently Mutated Cancer Genes and Tumor Mutation Burden in Chinese Breast Cancer. Front Oncol 2021;11:618767. [Crossref] [PubMed]
- Zelinova K, Jagelkova M, Laucekova Z, et al. Molecular analysis of circulating tumor DNA from breast cancer patients before and after surgery and following adjuvant chemotherapy. Mol Clin Oncol 2020;13:26. [Crossref] [PubMed]
- Li S, Lai H, Liu J, et al. Circulating Tumor DNA Predicts the Response and Prognosis in Patients With Early Breast Cancer Receiving Neoadjuvant Chemotherapy. JCO Precis Oncol 2020;4. ePO. [Crossref] [PubMed]
- Lu C, Dong XR, Zhao J, et al. Association of genetic and immuno-characteristics with clinical outcomes in patients with RET-rearranged non-small cell lung cancer: a retrospective multicenter study. J Hematol Oncol 2020;13:37. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213-9. [Crossref] [PubMed]
- Li J, Lupat R, Amarasinghe KC, et al. CONTRA: copy number analysis for targeted resequencing. Bioinformatics 2012;28:1307-13. [Crossref] [PubMed]
- Barroso-Sousa R, Jain E, Cohen O, et al. Prevalence and mutational determinants of high tumor mutation burden in breast cancer. Ann Oncol 2020;31:387-94. [Crossref] [PubMed]
- Kawaji H, Kubo M, Yamashita N, et al. Comprehensive molecular profiling broadens treatment options for breast cancer patients. Cancer Med 2021;10:529-39. [Crossref] [PubMed]
- Garcia-Murillas I, Chopra N, Comino-Méndez I, et al. Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol 2019;5:1473-8. [Crossref] [PubMed]
- Parsons HA, Rhoades J, Reed SC, et al. Sensitive Detection of Minimal Residual Disease in Patients Treated for Early-Stage Breast Cancer. Clin Cancer Res 2020;26:2556-64. [Crossref] [PubMed]
- O'Leary B, Cutts RJ, Liu Y, et al. The Genetic Landscape and Clonal Evolution of Breast Cancer Resistance to Palbociclib plus Fulvestrant in the PALOMA-3 Trial. Cancer Discov 2018;8:1390-403. [Crossref] [PubMed]
- Gao JJ, Swain SM, Luminal A. Breast Cancer and Molecular Assays: A Review. Oncologist 2018;23:556-65. [Crossref] [PubMed]
- Ellis H, Ma CX. PI3K Inhibitors in Breast Cancer Therapy. Curr Oncol Rep 2019;21:110. [Crossref] [PubMed]
- Saal LH, Holm K, Maurer M, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res 2005;65:2554-9. [Crossref] [PubMed]
- Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res 2008;68:6084-91. [Crossref] [PubMed]
- Kalinsky K, Jacks LM, Heguy A, et al. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res 2009;15:5049-59. [Crossref] [PubMed]
- André F, Ciruelos EM, Juric D, et al. Alpelisib plus fulvestrant for PIK3CA-mutated, hormone receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer: final overall survival results from SOLAR-1. Ann Oncol 2021;32:208-17. [Crossref] [PubMed]
- Mosele F, Stefanovska B, Lusque A, et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann Oncol 2020;31:377-86. [Crossref] [PubMed]
- Loi S, Sirtaine N, Piette F, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J Clin Oncol 2013;31:860-7. [Crossref] [PubMed]
- Rugo HS, Delord JP, Im SA, et al. Safety and Antitumor Activity of Pembrolizumab in Patients with Estrogen Receptor-Positive/Human Epidermal Growth Factor Receptor 2-Negative Advanced Breast Cancer. Clin Cancer Res 2018;24:2804-11. [Crossref] [PubMed]
- Ravaioli S, Limarzi F, Tumedei MM, et al. Are we ready to use TMB in breast cancer clinical practice? Cancer Immunol Immunother 2020;69:1943-5. [Crossref] [PubMed]
- Barroso-Sousa R, Keenan TE, Pernas S, et al. Tumor Mutational Burden and PTEN Alterations as Molecular Correlates of Response to PD-1/L1 Blockade in Metastatic Triple-Negative Breast Cancer. Clin Cancer Res 2020;26:2565-72. [Crossref] [PubMed]
- Wang S, Zhang Q, Yu C, et al. Immune cell infiltration-based signature for prognosis and immunogenomic analysis in breast cancer. Brief Bioinform 2021;22:2020-31. [Crossref] [PubMed]
- Sui S, An X, Xu C, et al. An immune cell infiltration-based immune score model predicts prognosis and chemotherapy effects in breast cancer. Theranostics 2020;10:11938-49. [Crossref] [PubMed]
- Jang BS, Han W, Kim IA. Tumor mutation burden, immune checkpoint crosstalk and radiosensitivity in single-cell RNA sequencing data of breast cancer. Radiother Oncol 2020;142:202-9. [Crossref] [PubMed]
- Chen K, Zhao H, Shi Y, et al. Perioperative Dynamic Changes in Circulating Tumor DNA in Patients with Lung Cancer (DYNAMIC). Clin Cancer Res 2019;25:7058-67. [Crossref] [PubMed]
- Ma F, Guan Y, Yi Z, et al. Assessing tumor heterogeneity using ctDNA to predict and monitor therapeutic response in metastatic breast cancer. Int J Cancer 2020;146:1359-68. [Crossref] [PubMed]
- Li H, Xu Y, Zhao F, et al. Plasma PIK3CA ctDNA specific mutation detected by next generation sequencing is associated with clinical outcome in advanced breast cancer. Am J Cancer Res 2018;8:1873-86. [PubMed]
- Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015;7:302ra133. [Crossref] [PubMed]
- Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, et al. Clinical significance of plasma cell-free DNA mutations in PIK3CA, AKT1, and ESR1 gene according to treatment lines in ER-positive breast cancer. Mol Cancer 2018;17:67. [Crossref] [PubMed]
- Yang J, Gong Y, Lam VK, et al. Deep sequencing of circulating tumor DNA detects molecular residual disease and predicts recurrence in gastric cancer. Cell Death Dis 2020;11:346. [Crossref] [PubMed]
(English Language Editor: J. Jones)



