
The expression of m6A regulators correlated with the immune microenvironment plays an important role in the prognosis of pancreatic ductal adenocarcinoma
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a malignant tumor of the digestive system that accounts for 95% of all pancreatic cancers (1). PDAC is considered to be a devastating malignant tumor because of its invasiveness, advanced stage and resistance to most treatments (2). Surgical resection remains the only radical treatment option; however, less than 20% of patients are eligible for resection (3). Compared with all other solid tumors, the 5-year survival rate of PDAC is about 9% (4); hence, it is urgent to discover genes that may influence the prognosis of patients with PDAC in clinical practice. At present, many studies have attempted to categorize PDAC patients; for example, by stratifying PDAC patients according to their genetic/epigenetic characteristics (5,6).
Several studies have revealed a series of DNA epigenetic changes in PDAC progression, and these findings have demonstrated significant clinical value for PDAC (7-10). Due to recent developments in technology, the modification of N6-methyladenosine (m6A) in mRNA has been reported. The m6A modification reveals a wide and rare external spectroscopic landscape, which has been found to be associated with a variety 3–4 of biological processes, including cancer (11). m6A introduction (methylation) and m6A removal (demethylation) may affect genes and pathways associated with the malignant phenotype of cancer (11). Nevertheless, as a new epigenetic modification, the role of RNA methylation in PDAC has not been well studied. At present, there are scant reports on the correlation between RNA methylation modification and PDAC (12,13). The m6A methylation is associated with high levels of expression of intracellular methyltransferase (“writer”) and demethylase (“eraser”), while the binding protein (“reader”) is bound to the m6A methylation site to execute multiple biological functions (14,15). As far as we know, there remains a lack of overall analysis of the expression of m6A methylation genes in PDAC with respect to clinical pathological characteristics, progression and prognosis.
A growing body of evidence supports a special correlation between m6A methylation regulatory genes and immune cells. YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) deficiency enhances the anti-tumor response of CD8+ T cells and strengthens the efficacy of anti-programmed death ligand 1 (PD-L1) therapy (16). AlkB homolog 5, RNA demethylase (ALKBH5) modulates the anti-programmed death 1 (PD-1) therapy response via regulating lactate and inhibiting immune cell accumulation in the tumor microenvironment (17). Methyltransferase 3, N6-adenosine-methyltransferase Complex Catalytic Subunit (METTL3) regulates T-cell homeostasis by modulating the interleukin (IL)-7/signal transducer and activator of transcription 5 (STAT5)/suppressors of cytokine signaling (SOCS) pathways (18). Nevertheless, the relationship between m6A regulators and immune cells in PDAC remains unknown. Thus, a global assessment of the immune landscape mediated by various m6A methylation regulators will help us to comprehensively understand the immune regulation of m6A regulators in the tumor immune microenvironment.
Compared with previous studies, this study is the first to comprehensively analyze the correlation between m6A methylation regulators and immune cells, prognosis, and clinicopathological features in PDAC. In this study, we established cluster subtype and risk model for m6A methylation regulators to improve prognostic risk stratification and facilitate treatment decisions in patients with PDAC. This study may provide a novel perspective for revealing the relationship between m6A methylation regulators and the tumor immune microenvironment in PDAC. We present the following article in accordance with the REMARK reporting checklist (available at https://gs.amegroups.com/article/view/10.21037/gs-21-859/rc).
Methods
Data processing
The gene expression data and full clinical information of PDAC were obtained from The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) database. As the main cohort, our study included gene expression and somatic mutation data of PDAC and normal pancreatic tissue samples in TCGA through the University of California Santa Cruz (UCSC) Xena platform (https://gdc.xenahubs.net). A total of 176 PDAC patients and 4 patients with normal pancreatic tissue were assigned into a training cohort. The datasets GSE28735, GSE62452, GSE71729 and GSE85916 were downloaded from the GEO database, and an averaging method using the RMA algorithm in the affy package (version 1.64.0, https://bioconductor.org/packages/affy/) was applied to execute background adjustment and quantile normalization. The ComBat, from the Surrogate Variable Analysis (sva) package (version 3.34.0, https://bioconductor.org/packages/sva/), was utilized to correct for batch effects. GSE28735 included 84 samples, including 42 PDAC tumor and 42 normal pancreatic tissue samples. GSE62452 contained 126 samples, including 65 tumor and 61 adjacent tissue samples. GSE71729 included 169 samples, including 123 PDAC tumor and 46 normal pancreatic tissue samples. There were only 79 PDAC tumor samples in GSE85916. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identification of m6A methylation regulatory genes
We found 19 m6A regulators in the published literature (11,14), including 2 erasers (ALKBH5 and FTO), 11 readers (IGF2BP1, IGF2BP2, IGF2BP3, LRPPRC, YTHDC1, YTHDC2, YTHDF1, YTHDF2, YTHDF3, FMR1, and HNRNPA2B1) and 6 writers (KIAA1429, METTL3, RBM15, RBM15B, WTAP, and ZC3H13). The mutation frequency of the m6A regulator was analyzed using R (version 3.5.3) software package (Bioconductor, https://www.bioconductor.org/). Due to the small number of paracancerous samples in the TCGA-PAAC dataset, we used the GSE28735, GSE62452, GSE71729, and GSE85916 datasets to compare the expression of m6A regulators in PDAC and control tissues. The difference in the expression of m6A regulators between PDAC and normal controls was calculated using the Wilcoxon signed rank test. A value of P<0.05 was deemed statistically significant.
Consensus clustering analysis of m6A methylation regulators
To reveal the biological characteristics of m6A methylation regulators in PDAC, the 176 PDAC patients in the TCGA- PDAC dataset were divided into subgroups by consensus clustering based on the expression of these m6A methylation regulators using the ConsensusClusterPlus R-package software (version 1.50.0, https://bioconductor.org/packages/ConsensusClusterPlus/), and the number of groups was shown by “k”. In order to uncover the correlation between consensus clustering and the clinicopathological features in PDAC, Pearson correlation analysis was applied to analyze the distribution of sex, cancer grade, cancer stage, survival state, cancer relapse and drink among the different clusters, and these results were displayed applying the pheatmap R-package software (version 1.0.12, https://cran.r-project.org/web/packages/pheatmap/index.html). The Kaplan-Meier method was used to evaluate survival curves and compare differences in survival among the different subgroups.
Comparison of immune cell infiltration among the m6A subgroups
The single sample gene set enrichment analysis (ssGSEA) algorithm was applied to study differences in immune cell infiltration between the different subgroups, and results were presented in a violin diagram. Stromal score, immune score, Estimation of Stromal and Immune cells in Malignant Tumor tissues using Expression data (ESTIMATE) score, and tumor purity were then evaluated by the ESTIMATE method according to TCGA-PDAC data. To explore the relationship between PD-L1 and these m6A regulators, the expression of PD-L1 in the two subtypes was evaluated using Pearson correlation analysis.
Gene set variation analysis (GSVA)
To study the effects of these m6A regulators on biological processes in different subgroups, GSVA was performed utilizing the GSVA package (version 1.34.0, https://bioconductor.org/packages/GSVA/) in R software. The predefined gene set “c2.cp.kegg.v7.2.symbols.gmt” was obtained from the Molecular Signatures Database (MSigDB) to execute the GSVA analysis. Then, the differentially expressed pathways were analyzed using the limma package in R software (version 3.42.2, https://bioconductor.org/packages/limma/). A false discovery rate (FDR) <0.01 was considered significant.
Construction of the m6A gene-related prognostic model
To better forecast the survival of PDAC patients, we conducted the m6A gene-related prognostic model according to the following steps. First, we performed a univariate Cox regression analysis to screen the m6A-related genes significantly associated with the prognosis of PDAC. For P<0.05, the gene was considered significant. Then, least absolute shrinkage and selection operator (LASSO)-Cox regression analysis was used to determine the prognostic model, and a 10-fold cross validation was used to ensure the optimal value of the LASSO penalty parameter. Based on the linear combination of the regression coefficient (β) of the LASSO-Cox regression model and the gene expression level, the m6A gene-related prognostic model was built. Finally, we developed the m6A gene-related prognostic model (5 genes) according to the following risk score formula:
where n represents the number of prognostic genes, expi represents the expression value of gene I, and βi represents the univariate Cox regression coefficient of gene i. Based on the median signature risk score as the cut-off point, PDAC patients in the TCGA-PDAC cohort were divided into low-risk and high-risk groups. To better evaluate the prognostic value of these 5 genes, we produced the survival curves for the high- and low-risk groups using the Kaplan-Meier method. In addition, a receiver operating characteristic (ROC) curve was produced to evaluate the accuracy and efficiency of these 5 genes in a time-dependent manner. Furthermore, the same risk scoring formula was run on the GSE28735, GSE62452, GSE71729, and GSE85916 datasets to further confirm the efficiency of these gene signatures. To ensure the predictive power of the prognostic model for PDAC patients was independent of other clinical variables, univariate and multivariate Cox regression analyses were used to analyze the prognostic gene signature and clinicopathological features in the TCGA-PDAC cohort.
Evaluation of the association between tumor risk score and tumor mutation burden (TMB)
Significant evidence suggests that the TMB may determine individual responses to cancer immunotherapy. Exploring the intrinsic link between TMB and risk score to clarify the genetic characteristics of each subgroup is of great significance. Corresponding information on the somatic alteration of the TCGA-PDAC cohort was obtained from the TCGA dataset. The maftools package in R software was used to calculate the number of somatic non-synonymous point mutations within each sample.
m6A modification patterns in the role of anti-programmed cell death ligand 1 (anti-PD-L1) immunotherapy
The immunotherapeutic advanced urothelial cancer (IMvigor210) cohort was included in our study to explore the predictive ability of the tumor risk score to assess the benefit of immunotherapy. Through the Creative Commons 3.0 License, the complete expression data and clinical annotations were obtained (http://research-pub.gene.com/IMvigor210CoreBiologies). The raw count data were normalized by the DEseq2 R-package software and then the count value was transformed into the transcripts-per-million (TPM) value.
Effect of immune cell infiltration-based m6A regulators on somatic copy number alterations (CNAs)
The effect of CNAs of the m6A regulators on immune cell infiltration levels was assessed using the Tumor Immune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/) which is composed of six immune cell types (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells).
Survival analysis
We assessed the effects of 5 m6A methylation regulators (ALKBH5, IGF2BP2, IGF2BP3, LRPPRC, and KIAA1429) on overall survival in patients with PDAC using clinical data from the TCGA-PDAC cohort. The log-rank test was used to test whether the differences in survival time were significant. Kaplan-Meier curves were visualized using the survival packages in R.
Immunohistochemistry analysis
The pictures of immunohistochemistry were obtained from The Human Protein Atlas (THPA; www.proteinatlas.org/), a publicly available database which includes more than 5 million images of immunohistochemically stained tissues and cells. Data regarding the expression levels of ALKBH5 and LRPPRC in PDAC tumor tissues were downloaded from THPA.
Gene Expression Profiling Interactive Analysis (GEPIA)
GEPIA is a newly developed interactive web server for analyzing the RNA sequencing expression data of 9,736 tumors and 8,587 normal samples from the TCGA and the Genotype-Tissue Expression projects, using a standard processing pipeline. The expression analysis of 5 m6A methylation regulators (ALKBH5, IGF2BP2, IGF2BP3, LRPPRC, and KIAA1429) was performed using GEPIA.
Statistical analysis
R software was used for all statistics analysis. The Wilcoxon test was used to calculate the significance of differences in immune cell infiltration and m6A methylation regulators expression. The survival curve was generated by Kaplan-Meier method, and the difference between groups was compared by log rank test. In addition, Pearson correlation analysis was performed. P<0.05 was considered statistically significant.
Results
Genetic alteration of the m6A regulators in patients with PDAC
On reviewing the literature, we found 19 m6A regulators, including 2 erasers, 11 readers, and 6 writers. We first summarized the incidence of somatic mutations in 19 m6A regulators in PDAC and identified the top 10 genes with the most frequent somatic mutations in PDAC (Figure 1A). Among 158 samples, the TP53 and Kirsten rat sarcoma virus (KRAS) genes showed the highest mutation frequencies. Only some of the 19 m6A methylation regulators showed low-mutation frequency.
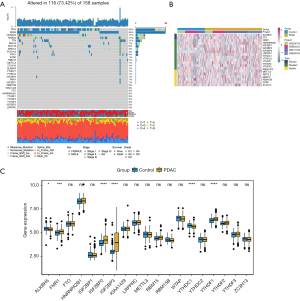
Identification of m6A methylation regulatory genes
We studied the expression of 19 m6A regulatory genes in PDAC and normal pancreatic tissues based on the GEO dataset. The clustering analysis of 19 m6A regulatory genes is shown in Figure 1B. The results showed that the expression level of 6 m6A regulation genes was significantly different between PDAC and normal tissues. The expression of FMR1, IGF2BP2, IGF2BP3, and YTHDC2 in PDAC tissues significantly increased, while the expression of ALKBH5 and YTHDC1 significantly decreased (Figure 1C).
Consensus clustering analysis of m6A methylation regulatory genes
From the gene expression data of TCGA-PDAC, we acquired the gene expression profiles of 176 PDAC patients. Following the similarity of expression of m6A methylation regulators in the PDAC samples, consensus clustering analysis was performed on these samples. In the process of consensus matrix K increasing from 2 to 5, when K=2, the crossover between the PDAC samples was the least (Figure 2A,2B); therefore, we applied consensus cluster K=2 to divide the PDAC patients into two subtypes, namely cluster A (n=71) and cluster B (n=105) (Figure 2C). In addition, we generated a boxplot (Figure 2D) for visualizing the expression of the 19 m6A regulators in cluster A and cluster B, and showed that the expression of ALKBH5, FTO, and RBM15 in cluster B was markedly lower than that in cluster A, while the expression of HNRNPA2B1, IGF2BP2, IGF2BP3, and LRPPRC was observably higher in cluster B than in cluster A. Further, the expression of individual m6A methylation regulators was higher in cluster B than in cluster A, especially the expression of IGF2BP2 and IGF2BP3. Furthermore, on examining the clinicopathological features in the two clusters, it was found that cluster A mainly involved female PDAC patients and was associated with high tumor differentiation (Figure 2E). Kaplan-Meier analysis showed that the overall survival time of cluster B was shorter than that of cluster A (Figure 3A). Finally, this was verified in the GEO datasets where similar results were obtained (Figure 3B). Our data showed that the clustering subtype defined by m6A methylation regulators expression was closely related to the heterogeneity of PDAC patients.

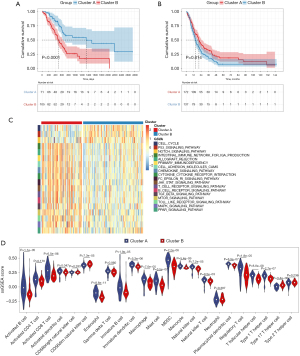
Comparison of immune cell infiltration among the m6A patterns
To uncover biological behavior between different PDAC subtypes, we executed GSVA and ssGSEA analysis. The results of GSVA showed that, in the cluster B subtype, immune-related pathways, such as the T-cell receptor signaling pathway and the B-cell receptor signaling pathway, were significantly inhibited, while other pathways related to PDAC progress were significantly increased, including the cell cycle and p53 signaling pathway (Figure 3C). ssGSEA results found that, in the cluster B subtype, the infiltration of immune cells in the tumor immune microenvironment was significantly reduced (Figure 3D); in particular, for activated B cells, activated CD8 T cells, immature B cells, and regulatory T cells. The results of the ESTIMATE algorithm also confirmed that, in the cluster B subtype, the Stromal Score, the Immune Score, and the ESTIMATE Score were observably lower than those scores of the cluster A subtype, and the tumor purity was dramatically higher than that of the cluster A subtype (Figure 4A). To reveal the relationship between PD-L1 and m6A methylation regulators, we assessed the expression of PD-L1 in two subtypes and found that the expression of PD-L1 in cluster B was observably higher than that in cluster A in both the TCGA cohort and the GEO datasets (Figure 4B,4C).
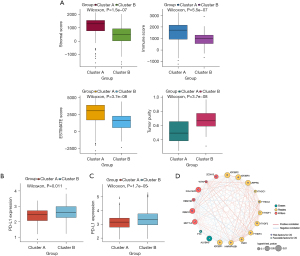
Establishment and verification of prognostic signatures
Based on the protein-protein interaction network analysis and the Pearson correlation analysis, we observed that the m6A methylation regulators had a cooperative relationship (Figure 4D). To further reveal the prognostic value of m6A methylation regulators in PDAC, the univariate Cox regression analysis was used to reveal the relationship between 19 m6A genes and the overall survival of PDAC patients in the TCGA cohort; 6 survival-related m6A methylation regulators (ALKBH5, IGF2BP2, IGF2BP3, LRPPRC, KIAA1429, and RBM15) were identified with P<0.01 as cut-off values (Figure 5A). Next, LASSO-Cox regression analysis for the 6 m6A methylation regulators was carried out to form a comprehensive and valid prognostic risk signature. We identified 5 m6A methylation regulators (ALKBH5, IGF2BP2, IGF2BP3, LRPPRC, and KIAA1429) to establish the risk model, and the coefficients of these 5 m6A methylation regulators were used to compute the risk score (Figure 5B,5C), as follows: Risk score = (0.3453× KIAA1429) + (0.3068× IGF2BP2) + (0.0704× IGF2BP3) + (0.0278× LRPPRC) − (0.4572× ALKBH5). Patients were divided into high-risk and low-risk groups with the median cutoff of risk score. Kaplan-Meier analysis illustrated that the overall survival time in the high-risk group was significantly shorter than that in the low-risk group (Figure 5D). The risk score distribution, survival status, and cluster analysis of these 5 prognostic m6A methylation regulators are presented in Figure 6A. In the high-risk group, the m6A high-risk regulatory factors LRPPRC, KIAA1429, IGF2BP2, and IGF2BP3 were highly expressed, while the protective m6A regulator ALKBH5 was up-regulated in the low-risk group. To better assess the prognostic accuracy of the signature composed of 5 m6A methylation regulators, we executed ROC analyses at 1, 3, and 5 years to compare their AUC values. The ROC results revealed that the AUC of the 1-, 3-, and 5-year plots were 0.66, 0.75, and 0.72, respectively, which indicated that the signature of 5 m6A methylation regulators provided a good discrimination performance for the prognosis of patients with PDAC (Figure 6B). Univariate analysis discovered that, applying the TNM-staging for PDAC, the T stage, the N stage, the TNM stage, the tumor grade, and the risk score were observably associated with overall survival of PDAC (Figure 6C). Moreover, multivariate Cox regression analysis found that the N stage and the risk score were markedly correlated with the prognosis of PDAC patients (Figure 6D). The above data showed that the risk score based on the signature of 5 m6A regulators was an independent prognostic factor for PDAC patients.

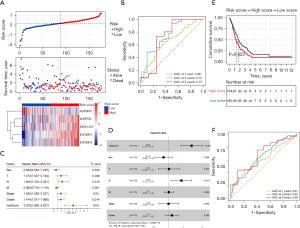
The GEO datasets were utilized to verify the prognostic value of the risk model. The same formula was used to calculate a risk score for each patient, and to classify patients into low-risk and high-risk groups based on the median risk score. The Kaplan-Meier analysis displayed that the overall survival time in the high-risk group was significantly shorter than that in the low-risk group (Figure 6E), which was consistent with the results in the TCGA cohort. Furthermore, the ROC curve analysis found that the risk score model had good predictive efficiency for 1-, 3-, and 5-year overall survival, and the AUC values were 0.61, 0.65, and 0.69, respectively (Figure 6F). In conclusion, these gene signatures displayed good discriminative performance in both the TCGA cohort and GEO datasets.
TMB in risk score
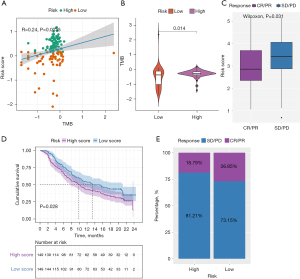
There is accumulating evidence that TMB is associated with responsiveness to immunotherapy. We studied the correlation between TMB and risk score and found that the risk score was positively correlated with the TMB (Figure 7A). Subsequently, the distribution patterns of TMB in the different risk score subgroups were revealed, and the TMB value was found to be higher in the high-risk score subgroup (Figure 7B).
m6A modification patterns in the role of anti-PD-L1 immunotherapy
Since there was no information about immunotherapy available in the TCGA-PDAC cohort, we further analyzed the response of immunotherapy. Subsequently, the IMvigor210 immunotherapy cohort was used to study whether the risk score could predict patients’ responses to anti-PD-L1 therapy. In the IMvigor210 cohort, patients with anti-PD-L1 immunotherapy were assigned to a high- or low-risk score group. Interestingly, as shown in Figure 7C, in the PD-L immune response group, in which the immune response was categorized as either a complete response (CR) or a partial response (PR), the risk score was significantly lower than that in the immune response-free group, in which the disease was categorized as either progressive disease (PD) or stable disease (SD). Furthermore, we found that patients classified as belonging to the low-risk group exhibited significant clinical survival benefits (Figure 7D). The frequencies of CR/PR and PD/SD were 26.85% and 73.15% in the low-risk patients, respectively, and 18.79% and 81.21% in the high-risk group patients, respectively (Figure 7E).
Effect of somatic CNAs of the m6A regulators on immune cell infiltration
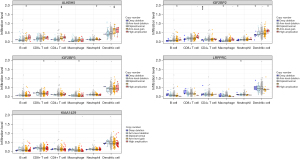
We further studied the effect of somatic CNAs of the 5 m6A regulators (ALKBH5, IGF2BP2, IGF2BP3, LRPPRC, and KIAA1429) on immune cell infiltration to clarify the potential mechanism of m6A methylation regulators in immune cell infiltration. As shown in Figure 8, the arm-level gain of ALKBH5 observably changed the infiltration levels of B cells, CD4+ T cells, neutrophils, and dendritic cells in PDAC. Furthermore, the CNAs of the IGF2BP2, including arm-level gain, deep deletion, arm-level deletion, and high amplification, markedly altered the infiltration levels of B cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in PDAC. This finding proved that these m6A regulators exerted pivotal regulatory effects on the tumor immune microenvironment for PDAC patients.
Gene and protein expression levels and survival analysis of the m6A methylation regulators
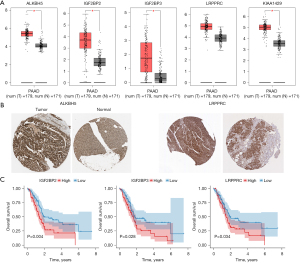
The expression analysis of the five m6A methylation regulators (ALKBH5, IGF2BP2, IGF2BP3, LRPPRC, and KIAA1429) was performed using GEPIA. We found that, except for LRPPRC, the other four methylation regulators (ALKBH5, IGF2BP2, IGF2BP3, and KIAA1429) were significantly upregulated in the tumor tissues of PDAC patients compared with the patients’ normal tissues (Figure 9A). The protein expression levels of ALKBH5 and LRPPRC were also significantly upregulated in the tumor tissue of PDAC patients compared with the patients’ normal tissues (Figure 9B). The results of the analysis of survival showed that only the expressions of IGF2BP2, IGF2BP3, and LRPPRC were significantly associated with survival in patients with PDAC (Figure 9C).
Discussion
Up to now, PDAC is still a fatal malignant tumor with a poor prognosis worldwide. The number of newly diagnosed cases and deaths of PDAC have observably increased at the same rate in recent years due to its poor prognosis (19). There is an urgent need to seek new biomarkers to predict the survival and monitor the treatment response for PDAC, and potentially provide new insights into the treatment of PDAC. Accumulative evidence has demonstrated that aberrant mRNA modification is strongly associated with the prognosis and overall survival of PDAC (20,21). m6A RNA methylation is essential for better predicting the malignant behavior and clinical prognosis of various cancers, and has attracted attention towards researching the prognosis of PDAC (22). Many studies have reported on the special relationship between m6A methylation regulators and the tumor immune microenvironment (23,24). However, this special relationship in PDAC has not been fully revealed; therefore, uncovering the correlation between m6A regulators and the tumor immune microenvironment will be helpful for the development of immune-based, targeted therapeutic strategies for PDAC.
In the current study, based on 19 m6A methylation regulators, we found two m6A modification patterns with dramatically different immune landscapes and prognoses. The degree of immune cell infiltration in TIME was significantly different between the two patterns. In cluster B with short survival time, the degree of immune cell infiltration in TIME was significantly reduced. This implies that the high degree of immune cell infiltration may be beneficial to the prognosis of PDAC. A prognostic gene signature of 5 m6A methylation regulators (ALKBH5, IGF2BP2, IGF2BP3, LRPPRC, and KIAA1429) was obtained using univariate Cox regression and LASSO Cox regression analysis, and the PDAC patients were divided into high-risk and low-risk groups based on the prognostic risk signature. Subsequently, we found that there were significant differences in the independent prognostic parameters, risk score distribution, survival status and cluster analysis between the high-risk and low-risk groups. Kaplan-Meier analysis illustrated that the overall survival time in the high-risk group was significantly shorter than that in the low-risk group. Moreover, the ROC results revealed that the AUCs of 1-, 3- and 5-year plots were 0.66, 0.75, and 0.72, respectively, which indicated that the signature of 5 m6A methylation regulators had a good discrimination performance for the prognosis of patients with PDAC. In addition, this gene signature displayed good discriminative performance in TCGA cohort and the GEO datasets. These results suggest that we can use the expression of 5 m6A methylation regulators to put patients into different groups, and then judge the prognostic characteristics of patients. The maftools package in R software was used to evaluate the association between the risk score and the TMB. We also found that the risk score was positively correlated with TMB, and the TMB value was higher in the high-risk group. However, the overall survival time in the high-risk group was significantly shorter than that in the low-risk group. Therefore, we hypothesized that TMB may reduce the survival time of PDAC. The IMvigor210 cohort was included in our study to explore the predictive ability of the risk score in determining the benefit of immunotherapy. Ultimately, we found that changes of CNAs in the somatic cells of m6A methylation regulators affected the infiltration of the immune cells, suggesting that these 5 m6A methylation regulators had a key regulatory effect on the tumor immune microenvironment of PDAC patients. Therefore, we hypothesized that 5 m6A methylation regulators may play an important role in the tumor immune microenvironment by regulating the infiltration degree of immune cells.
To illustrate the biological function in the two subtypes of PDAC patients, we carried out the GSVA enrichment analysis, and the results found that, compared with the cluster A subtype, immune-related pathways, such as the T cell receptor signaling pathway and the B cell receptor signaling pathway, were significantly inhibited in the cluster B subtype, while other pathways related to PDAC progress, such as the cell cycle and the p53 signaling pathway, were significantly increased. It is well known that the cell cycle and the p53 signaling pathway play important roles in the development and progression of PDAC (25-27). The results of this study further suggest that m6A methylation regulators may play an important role in the pathologic progression of PDAC by regulating related signaling pathways. In addition, the expression of PD-L1 in cluster B was significantly higher than that in cluster A in both TCGA cohort and the GEO datasets. Wang et al. have revealed that high expression of PD-L1 indicates a poor prognosis of PDAC (28). Lux et al. have reported that postoperative survival time of PDAC patients with high PD-L1 expression was observably shortened (29). Similarly, cluster B with high PD-L1 expression in our study had a shorter overall survival than cluster A with low PD-L1 expression, indicating that the consensus cluster analysis of m6A methylation regulatory genes performed in this study was reliable, which was consistent with the results of previous studies (28,29). PD-1 /PD-L1 can negatively regulate immune response and cause immune escape of tumor cells by inhibiting activation and proliferation of T cells. These results suggest that m6A methylation regulators may influence T cell activation and proliferation by regulating PD-L, thus mediating tumor immune tolerance.
Interestingly, we generated prognostic risk signatures based on 5 m6A methylation regulators (ALKBH5, IGF2BP2, IGF2BP3, LRPPRC, and KIAA1429) through univariate Cox regression and LASSO Cox regression analysis, which could be used to accurately predict the prognosis of PDAC. In addition, these 5 m6A methylation regulators were abnormally expressed in PDAC and correlated with prognosis of PDAC. Therefore, we speculated that these 5 m6A methylation regulators may be potential therapeutic targets for PDAC. ALKBH5, as a demethylation enzyme, is involved in methylation reversal. ALKBH5 is abnormally expressed in many cancers, including gastric cancer, glioblastomas, and epithelial ovarian cancer (30-32). ALKBH5 is a novel biomarker for predicting the prognosis of pancreatic cancer (33). ALKBH5 decreases in pancreatic cancer cells and regulates pancreatic cell motility through regulating KCNK15-AS1 demethylation (13). A decrease in ALKBH5 levels indicates that the clinical prognosis of PDAC is poor, and ALKBH5 overexpression suppresses the proliferation and metastasis of PDAC cells, while ALKBH5 silence is the opposite (34). Herein, ALKBH5 was significantly decreased between PDAC and normal tissues, and ALKBH5 was a key molecule in the consensus cluster and prognostic models of PDAC. Furthermore, the somatic CNAs of ALKBH5 observably changed the infiltration levels of B cells, CD4+ T cells, neutrophils, and dendritic cells in PDAC. Thus, the relationship of ALKBH5 with the level of immune cell infiltration in PDAC deserves further clarification.
Some studies have found that IGF2BP family acts as an m6A reader to regulate pancreatic cancer occurrence and progression. IGF2BP2 is upregulated in pancreatic cancer, and its expression is concerned with poor prognosis in pancreatic cancer patients (35,36). High expression of IGF2BP2 accelerates pancreatic cancer cell proliferation through regulating the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) signaling pathway (37). IGF2BP2 and IGF2BP3 are significantly increased in pancreatic cancer cells, and the inhibition of IGF2BP2 and IGF2BP3 can significantly reduce pancreatic cancer cell invasion (38). IGF2BP3 affects malignancy-related RNA regulation through modulating miRNA-mRNA interactions in PDAC (39). While in this study we also found that IGF2BP2 and IGF2BP3 were significantly upregulated in PDAC, the mechanism of their regulation of PDAC progress still needs to be further elucidated.
LRPPRC is found to be associated with mitochondrial translation and RNA decomposition, and its deficiency impacts the mitochondrial electron transport chain, mitochondrial permeability and transmembrane ROS diffusion (40,41). LRPPRC is inversely related to survival in pancreatic adenocarcinoma patients (42). KIAA1429 is a key component of the m6A methyltransferase complex. KIAA1429 promotes migration and invasion of hepatocellular carcinoma cell by impacting the m6A modification of ID2 mRNA (43). KIAA1429 promotes liver cancer progression via N6-methyladenosine-dependent GATA3 posttranscriptional modification (44). KIAA1429 regulates CDK1 as an oncogenic factor in breast cancer, in a manner independent of N6-methyladenosine (45). As far as we know, the function of KIAA1429 in PADC has not been studied. In the current study, we found that LRPPRC and KIAA1429 were key molecule in the consensus cluster and prognostic models of PDAC. Thence, the functions and potential mechanisms of LRPPRC and KIAA1429 in PDAC need to be further explored.
In conclusion, our study comprehensively studied the gene signatures of m6A-related regulators in PDAC. Two PDAC subtypes were obtained through the consensus clustering for m6A regulators; subsequently, the patients with PDAC were stratified, and showed significantly different prognoses and tumor-immune microenvironments. Moreover, the prognostic risk signatures constructed based on the expression levels of ALKBH5, IGF2BP2, IGF2BP3, LRPPRC, and KIAA1429, were not only closely correlated with clinical outcomes and clinicopathological features but were also independent prognostic indicators of PDAC. We also found that risk score was positively correlated with TMB, and the TMB value was higher in the high-risk group. The low-risk group was linked to a stronger response to anti-PD-L1 immunotherapy and clinical advantages in the IMvigor210 cohort. Furthermore, we correlated the somatic CNAs of these 5 m6A regulators with immune cell infiltration to clarify the potential mechanism of m6A methylation regulators related to different types of immune cell infiltration in PDAC. Specifically, the somatic CNAs of ALKBH5 and IGF2BP2 observably changed the infiltration levels of some immune cells. Our comprehensive assessment of m6A modification patterns in PDAC patients deepened our understanding of the tumor-immune landscape and provided a new approach for improving immunotherapy strategies in patients with PDAC. However, the conclusions of this study are based only on a bioinformatics analysis, therefore, prospective clinical studies are needed to verify the findings of this study.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://gs.amegroups.com/article/view/10.21037/gs-21-859/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://gs.amegroups.com/article/view/10.21037/gs-21-859/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Moffat GT, Epstein AS, O'Reilly EM. Pancreatic cancer-A disease in need: Optimizing and integrating supportive care. Cancer 2019;125:3927-35. [Crossref] [PubMed]
- Kamisawa T, Wood LD, Itoi T, et al. Pancreatic cancer. Lancet 2016;388:73-85. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Tan Z, Lei Y, Xu J, et al. The value of a metabolic reprogramming-related gene signature for pancreatic adenocarcinoma prognosis prediction. Aging (Albany NY) 2020;12:24228-41. [Crossref] [PubMed]
- Wu M, Li X, Liu R, et al. Development and validation of a metastasis-related Gene Signature for predicting the Overall Survival in patients with Pancreatic Ductal Adenocarcinoma. J Cancer 2020;11:6299-318. [Crossref] [PubMed]
- Majumder S, Raimondo M, Taylor WR, et al. Methylated DNA in Pancreatic Juice Distinguishes Patients With Pancreatic Cancer From Controls. Clin Gastroenterol Hepatol 2020;18:676-683.e3. [Crossref] [PubMed]
- Natale F, Vivo M, Falco G, et al. Deciphering DNA methylation signatures of pancreatic cancer and pancreatitis. Clin Epigenetics 2019;11:132. [Crossref] [PubMed]
- Dumitrescu RG. Early Epigenetic Markers for Precision Medicine. Methods Mol Biol 2018;1856:3-17. [Crossref] [PubMed]
- Chung M, Ruan M, Zhao N, et al. DNA methylation ageing clocks and pancreatic cancer risk: pooled analysis of three prospective nested case-control studies. Epigenetics 2021;16:1306-16. [Crossref] [PubMed]
- Wang S, Sun C, Li J, et al. Roles of RNA methylation by means of N6-methyladenosine (m6A) in human cancers. Cancer Lett 2017;408:112-20. [Crossref] [PubMed]
- Abukiwan A, Nwaeburu CC, Bauer N, et al. Dexamethasone-induced inhibition of miR-132 via methylation promotes TGF-β-driven progression of pancreatic cancer. Int J Oncol 2019;54:53-64. [PubMed]
- He Y, Hu H, Wang Y, et al. ALKBH5 Inhibits Pancreatic Cancer Motility by Decreasing Long Non-Coding RNA KCNK15-AS1 Methylation. Cell Physiol Biochem 2018;48:838-46. [Crossref] [PubMed]
- Yang Y. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res 2018;28:616-24. [Crossref] [PubMed]
- Yi L, Wu G, Guo L, et al. Comprehensive Analysis of the PD-L1 and Immune Infiltrates of m6A RNA Methylation Regulators in Head and Neck Squamous Cell Carcinoma. Mol Ther Nucleic Acids 2020;21:299-314. [Crossref] [PubMed]
- Han D, Liu J, Chen C, et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature 2019;566:270-4. Erratum in: Nature 2019;568:E3. [Crossref] [PubMed]
- Li N, Kang Y, Wang L, et al. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc Natl Acad Sci U S A 2020;117:20159-70. [Crossref] [PubMed]
- Li HB, Tong J, Zhu S, et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 2017;548:338-42. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Zhou Z, Zhang J, Xu C, et al. An integrated model of N6-methyladenosine regulators to predict tumor aggressiveness and immune evasion in pancreatic cancer. EBioMedicine 2021;65:103271. [Crossref] [PubMed]
- Tian J, Zhu Y, Rao M, et al. N6-methyladenosine mRNA methylation of PIK3CB regulates AKT signalling to promote PTEN-deficient pancreatic cancer progression. Gut 2020;69:2180-92. [Crossref] [PubMed]
- Hua YQ, Zhang K, Sheng J, et al. NUCB1 Suppresses Growth and Shows Additive Effects With Gemcitabine in Pancreatic Ductal Adenocarcinoma via the Unfolded Protein Response. Front Cell Dev Biol 2021;9:641836. [Crossref] [PubMed]
- Zhang B, Wu Q, Li B, et al. m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer 2020;19:53. [Crossref] [PubMed]
- Li Y, Gu J, Xu F, et al. Molecular characterization, biological function, tumor microenvironment association and clinical significance of m6A regulators in lung adenocarcinoma. Brief Bioinform 2021;22:bbaa225.
- Albahde MAH, Zhang P, Zhang Q, et al. Upregulated Expression of TUBA1C Predicts Poor Prognosis and Promotes Oncogenesis in Pancreatic Ductal Adenocarcinoma via Regulating the Cell Cycle. Front Oncol 2020;10:49. [Crossref] [PubMed]
- Hartman SJ, Bagby SM, Yacob BW, et al. WEE1 Inhibition in Combination With Targeted Agents and Standard Chemotherapy in Preclinical Models of Pancreatic Ductal Adenocarcinoma. Front Oncol 2021;11:642328. [Crossref] [PubMed]
- Kowalski S, Wyrzykowski D, Hac S, et al. New Oxidovanadium(IV) Coordination Complex Containing 2-Methylnitrilotriacetate Ligands Induces Cell Cycle Arrest and Autophagy in Human Pancreatic Ductal Adenocarcinoma Cell Lines. Int J Mol Sci 2019;20:261. [Crossref] [PubMed]
- Wang X, Li X, Wei X, et al. PD-L1 is a direct target of cancer-FOXP3 in pancreatic ductal adenocarcinoma (PDAC), and combined immunotherapy with antibodies against PD-L1 and CCL5 is effective in the treatment of PDAC. Signal Transduct Target Ther 2020;5:38. [Crossref] [PubMed]
- Lux A, Kahlert C, Grützmann R, et al. c-Met and PD-L1 on Circulating Exosomes as Diagnostic and Prognostic Markers for Pancreatic Cancer. Int J Mol Sci 2019;20:3305. [Crossref] [PubMed]
- Zhang J, Guo S, Piao HY, et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem 2019;75:379-89. [Crossref] [PubMed]
- Zhang S, Zhao BS, Zhou A, et al. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017;31:591-606.e6. [Crossref] [PubMed]
- Zhu H, Gan X, Jiang X, et al. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. J Exp Clin Cancer Res 2019;38:163. [Crossref] [PubMed]
- Cho SH, Ha M, Cho YH, et al. ALKBH5 gene is a novel biomarker that predicts the prognosis of pancreatic cancer: A retrospective multicohort study. Ann Hepatobiliary Pancreat Surg 2018;22:305-9. [Crossref] [PubMed]
- Tang B, Yang Y, Kang M, et al. m6A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Mol Cancer 2020;19:3. [Crossref] [PubMed]
- Hu X, Peng WX, Zhou H, et al. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ 2020;27:1782-94. [Crossref] [PubMed]
- Dahlem C, Barghash A, Puchas P, et al. The Insulin-Like Growth Factor 2 mRNA Binding Protein IMP2/IGF2BP2 is Overexpressed and Correlates with Poor Survival in Pancreatic Cancer. Int J Mol Sci 2019;20:3204. [Crossref] [PubMed]
- Xu X, Yu Y, Zong K, et al. Up-regulation of IGF2BP2 by multiple mechanisms in pancreatic cancer promotes cancer proliferation by activating the PI3K/Akt signaling pathway. J Exp Clin Cancer Res 2019;38:497. [Crossref] [PubMed]
- Cui XH, Hu SY, Zhu CF, et al. Expression and prognostic analyses of the insulin-like growth factor 2 mRNA binding protein family in human pancreatic cancer. BMC Cancer 2020;20:1160. [Crossref] [PubMed]
- Ennajdaoui H, Howard JM, Sterne-Weiler T, et al. IGF2BP3 Modulates the Interaction of Invasion-Associated Transcripts with RISC. Cell Rep 2016;15:1876-83. [Crossref] [PubMed]
- Cui J, Wang L, Ren X, et al. LRPPRC: A Multifunctional Protein Involved in Energy Metabolism and Human Disease. Front Physiol 2019;10:595. [Crossref] [PubMed]
- Cuillerier A, Honarmand S, Cadete VJJ, et al. Loss of hepatic LRPPRC alters mitochondrial bioenergetics, regulation of permeability transition and trans-membrane ROS diffusion. Hum Mol Genet 2017;26:3186-201. [Crossref] [PubMed]
- Hu S, Sechi M, Singh PK, et al. A Novel Redox Modulator Induces a GPX4-Mediated Cell Death That Is Dependent on Iron and Reactive Oxygen Species. J Med Chem 2020;63:9838-55. [Crossref] [PubMed]
- Cheng X, Li M, Rao X, et al. KIAA1429 regulates the migration and invasion of hepatocellular carcinoma by altering m6A modification of ID2 mRNA. Onco Targets Ther 2019;12:3421-8. [Crossref] [PubMed]
- Lan T, Li H, Zhang D, et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol Cancer 2019;18:186. [Crossref] [PubMed]
- Qian JY, Gao J, Sun X, et al. KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner. Oncogene 2019;38:6123-41. [Crossref] [PubMed]
(English Language Editor: K. Gilbert)


