
Prognostic value of tumor mutation burden and the relationship between tumor mutation burden and immune infiltration in HER2+ breast cancer: a gene expression-based study
Introduction
Human epidermal growth factor receptor-2 (HER2) positive breast cancer (BC) accounts for 15–20% of newly diagnosed invasive breast carcinomas (1). At present, anti-HER2 therapies, such as trastuzumab and pertuzumab, are the main treatment strategies (2). Moreover, HER2-targeting therapeutics have been approved, including drug-antibody conjugated ado-trastuzumab emtansine (T-DM1) and tyrosine kinase inhibitors (TKIs), such as lapatinib and pyrotinib (3-5). However, due to primary or secondary drug resistance against anti-HER2 therapies, some HER2+ BC patients will eventually progress (6). Thus, there is an urgent need to develop novel therapies and new approaches to overcome the limitations of targeted therapy and improve treatment.
Cancer immunotherapy, such as immune checkpoint blockade (ICB) with nivolumab, pembrolizumab and atezolizumab, has shown increasingly noteworthy value in the treatment of a variety of advanced and refractory cancers (7), such as in the treatment of melanoma (8) and lung cancer (9) and chimeric antigen receptor T cell therapy (ipilimumab) has been successfully utilized to treat refractory leukemia and lymphoma (10). Although BC is classified as a moderately immunogenic cancer (11,12), growing evidence suggests the existence of variable immunogenic activity in BC subtypes (13,14). Safonov et al. (15) found that increased immune metagene expression associated significantly with lower clonal heterogeneity in all subtypes of BC and with a trend for lower overall mutation, neoantigen, and CNV loads in TNBC and HER2+ cancers. While in estrogen receptor (ER)+ cancers, mutation load, neoantigen load, and CNV load weakly but positively associated with immune infiltration, which reached significance for overall mutation load only. Meanwhile, Schroth et al. (16) suggested that BRCA-related DNA repair deficiency and suppressed tumor immune responses may be clinically relevant predictors of endocrine therapy complementing treatment options in subgroups of hormone-sensitive early BC. Additionally, Musolino et al. (17) confirmed that the biological characteristics of Fc fragment of IgG receptor IIIa (FCGR3A) can facilitate the efficacy of trastuzumab by enhancing the antibody-dependent cell-mediated cytotoxicity (ADCC) effect. Therefore, based on the promising results from these studies, immunotherapy for HER2+ BC is a viable clinical objective.
Nevertheless, these immunotherapies are beneficial to only 20% of cancer patients (18). Thus, many efforts have been devoted to identifying more immunotherapy targets and clarifying molecular mechanism of immunotherapy responsiveness (19). Currently, programmed cell death-1/programmed cell death-ligand 1 (PD-1/PD-L1) expression on tumors (20), DNA mismatch-repair deficiency, neoantigen load (21), tumor-infiltrating lymphocytes (22) and IFN-γ-related mRNA profile (23) are recognized molecular determinants. However, a preliminary exploratory study has shown that monotherapy with PD-1/PD-L1 inhibitors is not satisfactory for BC, and the population that benefits from this therapy is limited (24). Tumor mutation burden (TMB) is also a potential biomarker for immunotherapy. Recent studies have found that TMB has a close relationship with immune infiltration and the prognosis of various cancers (25) and the TAPUR study suggested that BC patients in the high-TMB group can benefit from pembrolizumab monotherapy (26). However, the relationship between TMB-related genomic biomarkers and immune infiltration in HER2+ BC are still unclear. In this study, we analyzed the somatic mutations and RNA-seq data for HER2+ BC in TCGA and divided the samples into high- and low-TMB expression groups. Next, we analyzed the influence of TMB-related differentially expressed genes (DEGs) on the immune microenvironment. Finally, we established a risk model combining immune-related genes and mutant genes to evaluate prognosis for HER2+ BC patients. We believe that the risk score system have potential in patient management and that the selected hub genes have the potential to be combined targets for immunotherapy for HER2+ BC patients. We present the following article in accordance with the REMARK reporting checklist (available at https://gs.amegroups.com/article/view/10.21037/gs-21-594/rc).
Methods
Acquisition of somatic mutation and RNA-seq data
We prepared somatic mutation data for 216 HER2+ BC from the “Masked Somatic Mutation” category in TCGA database via the GDC data portal (https://portal.gdc.cancer.gov/). The “maftools” R package provides multiple analysis modules to perform the visualization process (27). Additionally, the corresponding clinical data sheets for HER2+ BC were obtained from cBioPortal (https://www.cbioportal.org/) and used as the training dataset. Of 114 HER2+ samples from GSE50948 and 323 HER2+ samples from GSE96058 in the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) were used as the validation datasets. In the GSE50948 dataset, we found that only 63 patients who had received a combination of trastuzumab and chemotherapy (CT) in neoadjuvant therapy, while the other patients received CT alone. Patient inclusion criteria: immunohistochemistry (IHC) or fluorescence in situ hybridization (FISH) was used to determine the status of HER2 in BC. If one of these was positive, we define these samples as HER2+ regardless of ER or progesterone receptor (PR) status. Patient exclusion criteria: (I) both IHC and FISH were negative or uncertain HER2 status; (II) patients with incomplete clinicopathological information, such as TX stage (the primary tumor could not be assessed), NX stage (regional lymph node involvement could not be assessed), and MX stage (the metastatic status could not be assessed) in the TNM staging system. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Calculation and grouping of TMB
We defined TMB as the total number of somatic mutations per coding area of a tumor genome or the total number of mutations per megabase (mut/Mb). Perl software was applied to calculate TMB, and only mutations that caused amino acid changes (nonsynonymous mutation) were included. In our cohort, the average TMB (3.05 mut/Mb) was approximately equivalent to the 80th percentile of the TMB distribution. Using the average TMB as the cutoff value, patients were divided into high- (n=43) and low-TMB (n=173) groups. Previous studies had shown that it is feasible to use the average as an initial threshold and verified it as an acceptable threshold choice by alternative cutpoint testing (28).
Prognostic analysis
Kaplan-Meier analyses with log-rank tests were subsequently performed to assess the differential outcomes in overall survival (OS) and disease-free survival (DFS) between the two levels by the “survival” R package. The definition of DFS: in cancer, the length of time after primary treatment for a cancer ends that the patient survives without any signs or symptoms of that cancer. Also called DFS, relapse-free survival, and RFS. In addition, we further evaluated the relationship between the TMB level and multiple clinical variables.
ImmuCellAI, TIMER and Immport database
ImmuCellAI (http://bioinfo.life.hust.edu.cn/ImmuCellAI#!/) is a tool that estimates the abundance of 24 types of immune cell infiltrates from a gene expression dataset (29). Based on the “Gene” and “sCNA” modules of the TIMER database (https://cistrome.shinyapps.io/timer/), we further evaluated the correlation between hub genes and immune microenvironment in HER2+ BCs. Moreover, we obtained 1,811 immune-related genes from the Immport database (https://www.immport.org/shared/home).
DEGs and functional analysis
The “edgeR” R package was utilized to identify DEGs between the two groups with |log (fold change) >1| and FDR <0.05. The “Venn” diagram identified 29 immune-related DEGs. We performed Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis on DEGs using the David database (https://david.ncifcrf.gov/). The difference between high- and low-TMB cohorts can be detected by the “maftools” R package, which performs a Fisher test on all genes to detect differentially mutant genes. We selected the first 30 mutant genes with significant differences.
Construction and validation of the immune- and mutant-related risk scoring system
After excluding patients with missing mutation information and distant metastasis, 178 samples were subjected to subsequent analysis. The clinical characteristics of all patients are shown in Table 1. In the immune- and mutant-related genes identified above, we finally obtained 17 immune genes and 6 mutant genes with prognostic significance in the Kaplan-Meier plotter database (https://kmplot.com/analysis/) (30). We used the Cox proportional hazard regression analysis (Cox regression analysis) to construct a risk scoring system and multiplied by the gene expression of each patient. A receiver operating characteristic (ROC) curve was drawn to assess the predictive power of the system using the “survivalROC” R package, and a Kaplan-Meier analysis with log-rank test was performed using the “survminer” R package.
Table 1
| Variables | TCGA cohort (n=178) | |
|---|---|---|
| n | % | |
| Status | 153 | 85.96% |
| Alive | 153 | 85.96% |
| Dead | 25 | 14.04% |
| Age, years | ||
| ≥60 | 81 | 45.51% |
| <60 | 97 | 54.49% |
| AJCC-T | ||
| T1 | 36 | 20.22% |
| T2 | 117 | 65.73% |
| T3 | 15 | 8.43% |
| T4 | 10 | 5.62% |
| AJCC-N | ||
| N0 | 74 | 41.57% |
| N1 | 68 | 38.20% |
| N2 | 25 | 14.04% |
| N3 | 11 | 6.18% |
| AJCC-M | ||
| M0 | 174 | 97.75% |
| M1 | 4 | 2.25% |
| Stage | ||
| Stage I | 20 | 11.24% |
| Stage II | 109 | 61.24% |
| Stage III | 45 | 25.28% |
| Stage IV | 4 | 2.25% |
| ER status | ||
| Positive | 123 | 69.10% |
| Negative | 55 | 30.90% |
| PR status | ||
| Positive | 95 | 53.37% |
| Negative | 83 | 46.63% |
BC, breast cancer; TCGA, The Cancer Genome Atlas; ER, estrogen receptor; PR, progesterone receptor.
Statistical analysis
The “Limma” package was utilized to conduct the normalization and differential analysis. The Wilcoxon rank-sum test was a nonparametric statistical test mainly utilized for comparing two groups, and the Kruskal-Wallis test was suitable for comparing two or more groups. The Chi-square test was used to compare composition ratios or rates between the two groups. All statistical analysis was implemented based on R software (Version 4.0.2, https://www.r-project.org/). The conventional two-sided P value <0.05 was considered significant.
Results
Landscape of the HER2+ BC mutation profiles
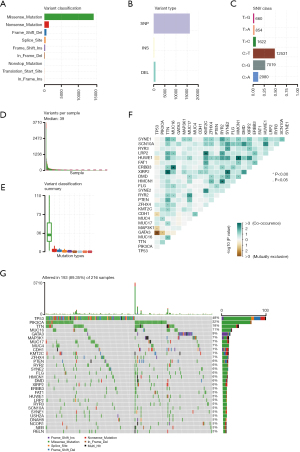
In summary, various mutation categories were categorized into different groups, in which missense mutations were the most common type, single nucleotide polymorphisms (SNPs) occurred more frequently than insertions or deletions, and C > T was the most common single nucleotide variant (SNV) (Figure 1A-1C). Moreover, we calculated the number of mutant bases in each sample and displayed the mutation categories using different colors in box plots (Figure 1D,1E). The coincident and exclusive associations were shown across mutant genes (Figure 1F). Mutation information for each gene in each sample is shown in a waterfall plot, and tumor protein P53 (TP53; 48%), phosphoinositide 3-kinase alpha (PIK3CA; 32%) and titin (TTN; 18%) were the top three mutant genes in the samples (Figure 1G).
TMB is correlated with survival, hormone receptor and immune infiltration of HER2+ BC
Using the average TMB as the cutoff point, we divided all samples into high- and low-TMB groups (Figure 2A). A higher level of TMB correlated with worse OS (P=0.008) and conferred a higher risk of tumor recurrence (P=0.03) (Figure 2B,2C). Moreover, a high TMB level was significantly related to hormone receptor negative status and advanced age (Figure 2D-2F). Nevertheless, no significant difference was found in TNM stages or pathological stages (Figure S1).
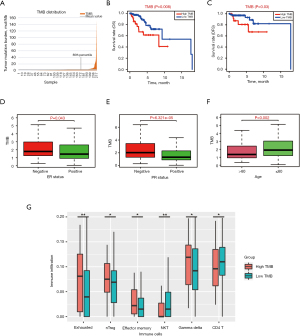
Based on the ImmuCellAI database, we estimated the abundance of 24 immune cells in each patient. In the high-TMB group, the infiltration levels of exhausted T cells, nature T regulatory cells (nTreg), effector memory T cells and gamma delta T cells were higher than that of the low-TMB group, while CD4+ T cells and Natural Killer (NK) cells were the opposite (Figure 2G). However, no significant differences were observed in the infiltration levels of other immune cells between the two TMB groups (Figure S2).
Identification of immune- and mutant-related DEGs between high- and low-TMB groups
A total of 182 DEGs (Table S1) were identified and are shown in a volcano plot (Figure 3A). We screened 29 immune-related DEGs in 1,811 immune genes from the Immport database by the “Venn” diagram (Figure 3B) and further identified 17 genes with prognostic significance in the Kaplan-Meier plotter database (Figure S3). In the biological process group, some immune processes involved in the functional analysis were the response to cytokines and the negative regulation of leukocyte differentiation (Figure 3C). KEGG pathway analysis indicated that these DEGs were mainly involved in the Rap1 signaling pathway (Figure 3D).
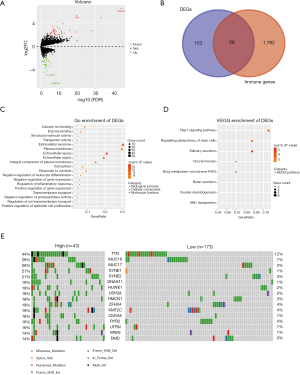
“Co-oncoplot” was used to map the two TMB groups side by side to better compare the differentially mutant genes, with the frequency and type of mutations shown (Figure 3E). We selected the first 30 of the 91 differentially mutant genes (Table S2) for prognostic analysis of mutation and expression, and finally, 6 mutant genes were identified (Figure S4).
Construction and validation of the immune- and mutant-related risk scoring system
After identifying 17 immune- and 6 mutant-related genes, we used the Cox regression analysis to construct an immune- and mutant-related risk scoring system including 5 hub genes. We performed univariate Cox regression analysis on each of the characteristics and for factors with a P value <0.10 (Table S3), then we further constructed the final risk model by multivariate Cox regression analysis. The risk scores formula is as follows: risk score =0.860788 × expDNAH11 + 0.505288 × expERBB3 − 0.41573 × expFABP5 − 0.43514 × expMAPT − 0.60184 × expPGR (Table 2). Then, the risk score of each sample was calculated. Patients in the high-risk group had worse survival outcomes (P=2.205e-04) (Figure 4A). The ROC curve of 5-year OS prediction was drawn to assess the predictive accuracy with area under curve (AUC) =0.759 (Figure 4B). Moreover, we also use 17 immune-related genes and 6 mutant-related genes to establish risk models, respectively (Figure S5A,S5B, Tables S4,S5). Altogether, the combined model was indeed better than the above two alone.
Table 2
| Genes | Coef | HR | HR.95L | HR.95H | P value |
|---|---|---|---|---|---|
| DNAH11 | 0.860788 | 2.365025 | 1.191053 | 4.696132 | 0.013912 |
| ERBB3 | 0.505288 | 1.657463 | 0.992778 | 2.767167 | 0.053329 |
| FABP5 | −0.41573 | 0.659858 | 0.379027 | 1.148763 | 0.141647 |
| MAPT | −0.43514 | 0.647174 | 0.45371 | 0.923132 | 0.016334 |
| PGR | −0.60184 | 0.547805 | 0.258784 | 1.159615 | 0.115735 |
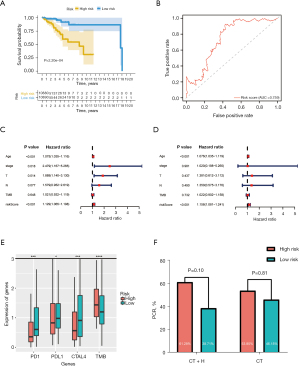
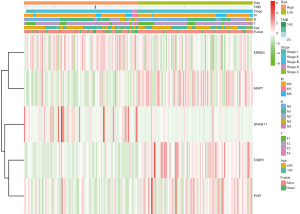
We utilized the univariate and multivariate Cox analysis to investigate the relationship of the risk score and clinical factors. Multivariate Cox analysis suggested that age and risk score are independent prognostic factors, but TMB was not (Figure 4C,4D). In addition, higher risk was related to higher TMB and lower PD-1, PD-L1, and CTLA-4 (Figure 4E). After grouping by risk value in the GSE50948 dataset, we found only 63 patients who had received a combination of trastuzumab and CT in neoadjuvant therapy, The pathologic complete response (pCR) of the high-risk group was 61.29%, while that of low-risk group was 38.7%. Moreover, the pCR was 53.85% in the high-risk group and 46.15% in the low-risk group for those who received CT alone (Figure 4F). The heat map shows the expression of the 5 hub genes and clinicopathological variables in the high- and low-risk groups (Figure 5). We verified that the expression level of ERBB3, DNAH11 in high-risk group was higher than low-risk group, while MAPT, FABP5, PGR were the opposite.
To verify that the system has good prognostic prediction capabilities, we screened out 323 HER2+ BC from the GSE96058 dataset and then applied the same risk scoring formula. High-risk patients had significantly worse prognosis (P=2.417e-02). ROC analysis showed that the system has a relatively good prognostic prediction (AUC =0.609) (Figure S5C).
Immune infiltration between the low- and high-risk groups
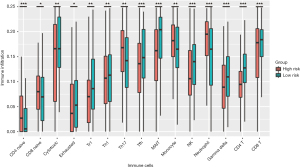
We explored the potential relationship between our risk scoring system and immune infiltration using the “ImmuCellAI” tool. Most of the immune cells in the high-risk group had a lower degree of infiltration, including CD4+ T cells, CD8+ T cells, NK cells, exhausted T cells, T regulatory type 1 cells (Tr1), follicular helper T cells (Tfh), mucosal-associated invariant T cells (MAIT), gamma delta T cells and cytotoxic T cells (Tc), in which CD4+ T and NK cells were consistent with the previous results of immune infiltration between high- and low-TMB groups. However, the higher infiltration degree of monocyte, neutrophil and helper T 17 cells (Th17) were observed in the high-risk group (Figure 6).
Based on the TIMER database, we further found that various forms of mutations carried by hub genes can usually suppress immune infiltration, including CD4+ and CD8+ T cells, neutrophils, macrophages, dendritic cells and NK cells (Figure S6). However, the expression of DNAH11 was negatively correlated with the infiltration level of immune cells, and the expression of other genes (except ERBB3) was positively correlated (Figure S7).
Discussion
In this study, we comprehensively studied the role of TMB in the regulation of the immune phenotype in HER2+ BC. Then, a risk scoring system related to immunity and mutation was developed based on the TCGA dataset. This risk scoring system has favorable prognostic prediction ability, which is independent of traditional prognostic factors. Furthermore, we explored the correlation between the risk score and immune cell infiltration in HER2+ BC. To our knowledge, this is the first prognostic model combining immune genes with mutant genes to predict survival outcomes in HER2+ BC.
TMB has been described as a predictor of tumor behavior and immunological response (31-33), and it has been demonstrated to be effective in a variety of tumors, such as colorectal cancer (34), lung cancer (35,36), etc. In our study, higher TMB was significantly correlated with worse OS and DFS. However, a recent bioinformatics study demonstrated that in HER2+ refractory metastatic BC, patients with lower TMB had worse survival outcome than those who had higher TMB (44.9 vs. 85.8 months, P=0.016) (37). The difference between the two studies is that the patients of the two studies are two independent cohorts. Samples for our study were derived from TCGA dataset, and patients at advanced stages were excluded. In addition, current studies have shown that trastuzumab can change the immune status of HER2+ BC by enhancing the ADCC effect, making patients more likely to benefit from this treatment (17,38). Only a small number of patients in our cohort received trastuzumab treatment. Thus, the treatment may lead to different levels of TMB expression and different prognosis. Our results indicated that TMB is a potential biomarker of immune-mediated survival in HER2+ BC.
Then, we identified five hub genes, including two mutant genes (DNAH11, ERBB3) and three immune-related genes (FABP5, MAPT, PGR), that were highly associated with OS in HER2+ BC. Then, a predictive model was developed. Patients with a high-risk score had poor survival outcomes, and the AUC of the ROC curve was 0.759, indicating the superior predictive accuracy. In our study, there were no mutations in differentially expressed immune-related genes. Therefore, we used Cox regression analysis to perform immune-related gene modeling (AUC =0.741) and mutant gene modeling (AUC =0.624). The result indicated that the prognosis modeled by only one class of genes was not as good as combination modeling. In short, our research proposed a combination modeling to predict the prognosis in HER2+ BC and requires validation in the future.
Furthermore, we analyzed the association between the risk score and the expression of critical immune checkpoint genes. The results showed that patients in the low-risk group had higher PD-1, PD-L1 and CTLA-4 expression. Thus, the patients in the low-risk group were more likely to benefit from ICBs. However, these results did not match with the low-TMB expression in the low-risk group. At present, the expression level of PD-L1 can be evaluated by immunohistochemistry (IHC), but there are still limitations to using PD-L1 as a biomarker of immunotherapy response. PD-L1 expression in tumors is heterogeneous and is affected by previous treatments, such as CT and radiotherapy (39,40). Furthermore, in the KEYNOTE-055 trial, researchers found that PD-L1-negative patients also had a significant response to pembrolizumab treatment, suggesting that evaluation of the immunotherapy response with PD-L1 alone is not sufficient (41). Importantly, some studies have also demonstrated that TMB has no association with PD-L1 expression, indicating that TMB and PD-L1 expression were independent biomarkers for predicting the response to ICBs (42,43). Some studies now showed that trastuzumab in combination with ICBs may benefit some patients. Su et al. (44) confirmed that trastuzumab can enhance the expression of PD-L1. The PANACEA study estimated that in the PD-L1-positive subgroup, the objective response rate (ORR) of trastuzumab combined with pembrolizumab was 15% (45); similarly, the KATE2 study showed that the 1-year OS rate of patients in the atezolizumab group was better (46). However, our study suggested that patients in the high-risk group were able to achieve a higher pCR trend with trastuzumab plus CT, and this is inconsistent with poorer outcomes in the high-risk group. Our results may indicate that patients in the high-risk group are more sensitive to CT plus trastuzumab, which may be not related to the treatment of trastuzumab, but be associated with the change of immune microenvironment. pCR rate is not the only prognostic factors, as clinical trial of NeoALTTO, for example, suggested no difference in OS and DFS between patients in the pCR group and those in the non-PCR group (47). Thus, more studies are investigating changes in immune markers after trastuzumab treatment to explore more methods and strategies for combined therapy. In terms of immune cell infiltration, more regulatory T cells and fewer NK cells were found in the high-TMB/high-risk group, consistent with similar results in another study of HER2+ BC, where more Treg cells and fewer NK cells demonstrated a worse prognosis (48). In summary, our study systematically explored the relationships between TMB and immune infiltration and constructed a TMB-related risk scoring system in HER2+ BC. Thus, our study provides new insights into the immune cell infiltration of the tumor microenvironment and immunotherapies for HER2+ BC.
There were some limitations to our study. First, this is a retrospective study, and the results should be further confirmed by prospective studies to exclude sample selection bias and inter-tumor heterogeneity. Additionally, the clinical relevance of the 5 hub genes need more independent validation in better-documented clinical trials in the future.
In conclusion, HER2+ BC patients with low TMB have a better prognosis than patients with high TMB. TMB could regulate immune cell infiltration. The immune- and mutant-related risk scoring system can divide patients into low- and high-risk groups with different outcomes and immunophenotypes.
Conclusions
Our study suggested that TMB is associated with poor prognosis of HER2 positive BC, and established a prognostic model for the combination of mutation-related genes and immune genes. Moreover, our results suggested that high-risk group are more likely to benefit from ICIs treatment, which provide a new strategy for evaluating the efficacy of immunotherapy and a new insight for finding HER2 positive BC patients who are not sensitive to immunotherapy.
Acknowledgments
Funding: This work was supported by grants from the Natural Science Foundation of the Hunan Province of China (2020JJ4828) and the Science and Technology Innovation Program of Hunan Province (2021SK2026).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://gs.amegroups.com/article/view/10.21037/gs-21-594/rc
Peer Review File: Available at https://gs.amegroups.com/article/view/10.21037/gs-21-594/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://gs.amegroups.com/article/view/10.21037/gs-21-594/coif). All authors report that this work was supported by the Natural Science Foundation of the Hunan Province of China (2020JJ4828) and the Science and Technology Innovation Program of Hunan Province (2021SK2026). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen X, Yuan Y, Gu Z, et al. Accuracy of estrogen receptor, progesterone receptor, and HER2 status between core needle and open excision biopsy in breast cancer: a meta-analysis. Breast Cancer Res Treat 2012;134:957-67. [Crossref] [PubMed]
- Swain SM, Kim SB, Cortés J, et al. Pertuzumab, trastuzumab, and docetaxel for HER2-positive metastatic breast cancer (CLEOPATRA study): overall survival results from a randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol 2013;14:461-71. [Crossref] [PubMed]
- Chen Q, Ouyang D, Anwar M, et al. Effectiveness and Safety of Pyrotinib, and Association of Biomarker With Progression-Free Survival in Patients With HER2-Positive Metastatic Breast Cancer: A Real-World, Multicentre Analysis. Front Oncol 2020;10:811. [Crossref] [PubMed]
- Ouyang DJ, Chen QT, Anwar M, et al. The Efficacy of Pyrotinib as a Third- or Higher-Line Treatment in HER2-Positive Metastatic Breast Cancer Patients Exposed to Lapatinib Compared to Lapatinib-Naive Patients: A Real-World Study. Front Pharmacol 2021;12:682568. [Crossref] [PubMed]
- Anwar M, Chen Q, Ouyang D, et al. Pyrotinib Treatment in Patients With HER2-positive Metastatic Breast Cancer and Brain Metastasis: Exploratory Final Analysis of Real-World, Multicenter Data. Clin Cancer Res 2021;27:4634-41. [Crossref] [PubMed]
- Chung A, Cui X, Audeh W, et al. Current status of anti-human epidermal growth factor receptor 2 therapies: predicting and overcoming herceptin resistance. Clin Breast Cancer 2013;13:223-32. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Pollack MH, Betof A, Dearden H, et al. Safety of resuming anti-PD-1 in patients with immune-related adverse events (irAEs) during combined anti-CTLA-4 and anti-PD1 in metastatic melanoma. Ann Oncol 2018;29:250-5. [Crossref] [PubMed]
- Gong X, Li X, Jiang T, et al. Combined Radiotherapy and Anti-PD-L1 Antibody Synergistically Enhances Antitumor Effect in Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:1085-97. [Crossref] [PubMed]
- June CH, O'Connor RS, Kawalekar OU, et al. CAR T cell immunotherapy for human cancer. Science 2018;359:1361-5. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. [Crossref] [PubMed]
- Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011;121:2750-67. [Crossref] [PubMed]
- Miller LD, Chou JA, Black MA, et al. Immunogenic Subtypes of Breast Cancer Delineated by Gene Classifiers of Immune Responsiveness. Cancer Immunol Res 2016;4:600-10. [Crossref] [PubMed]
- Safonov A, Jiang T, Bianchini G, et al. Immune Gene Expression Is Associated with Genomic Aberrations in Breast Cancer. Cancer Res 2017;77:3317-24. [Crossref] [PubMed]
- Schroth W, Büttner FA, Kandabarau S, et al. Gene Expression Signatures of BRCAness and Tumor Inflammation Define Subgroups of Early-Stage Hormone Receptor-Positive Breast Cancer Patients. Clin Cancer Res 2020;26:6523-34. [Crossref] [PubMed]
- Musolino A, Naldi N, Bortesi B, et al. Immunoglobulin G fragment C receptor polymorphisms and clinical efficacy of trastuzumab-based therapy in patients with HER-2/neu-positive metastatic breast cancer. J Clin Oncol 2008;26:1789-96. [Crossref] [PubMed]
- Braun DA, Burke KP, Van Allen EM. Genomic Approaches to Understanding Response and Resistance to Immunotherapy. Clin Cancer Res 2016;22:5642-50. [Crossref] [PubMed]
- Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol 2016;17:e542-51. [Crossref] [PubMed]
- Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther 2015;14:847-56. [Crossref] [PubMed]
- Efremova M, Finotello F, Rieder D, et al. Neoantigens Generated by Individual Mutations and Their Role in Cancer Immunity and Immunotherapy. Front Immunol 2017;8:1679. [Crossref] [PubMed]
- Zito Marino F, Ascierto PA, Rossi G, et al. Are tumor-infiltrating lymphocytes protagonists or background actors in patient selection for cancer immunotherapy? Expert Opin Biol Ther 2017;17:735-46. [Crossref] [PubMed]
- Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930-40. [Crossref] [PubMed]
- Nanda R, Chow LQ, Dees EC, et al. Pembrolizumab in Patients With Advanced Triple-Negative Breast Cancer: Phase Ib KEYNOTE-012 Study. J Clin Oncol 2016;34:2460-7. [Crossref] [PubMed]
- Wang X, Li M. Correlate tumor mutation burden with immune signatures in human cancers. BMC Immunol 2019;20:4. [Crossref] [PubMed]
- Alva AS, Mangat PK, Garrett-Mayer E, et al. Pembrolizumab in Patients With Metastatic Breast Cancer With High Tumor Mutational Burden: Results From the Targeted Agent and Profiling Utilization Registry (TAPUR) Study. J Clin Oncol 2021;39:2443-51. [Crossref] [PubMed]
- Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747-56. [Crossref] [PubMed]
- Thomas A, Routh ED, Pullikuth A, et al. Tumor mutational burden is a determinant of immune-mediated survival in breast cancer. Oncoimmunology 2018;7:e1490854. [Crossref] [PubMed]
- Miao YR, Zhang Q, Lei Q, et al. ImmuCellAI: A Unique Method for Comprehensive T-Cell Subsets Abundance Prediction and its Application in Cancer Immunotherapy. Adv Sci (Weinh) 2020;7:1902880. [Crossref] [PubMed]
- Lánczky A, Győrffy B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J Med Internet Res 2021;23:e27633. [Crossref] [PubMed]
- Goodman AM, Kato S, Bazhenova L, et al. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther 2017;16:2598-608. [Crossref] [PubMed]
- Birkbak NJ, Kochupurakkal B, Izarzugaza JM, et al. Tumor mutation burden forecasts outcome in ovarian cancer with BRCA1 or BRCA2 mutations. PLoS One 2013;8:e80023. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Schrock AB, Ouyang C, Sandhu J, et al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann Oncol 2019;30:1096-103. [Crossref] [PubMed]
- Hellmann MD, Callahan MK, Awad MM, et al. Tumor Mutational Burden and Efficacy of Nivolumab Monotherapy and in Combination with Ipilimumab in Small-Cell Lung Cancer. Cancer Cell 2018;33:853-861.e4. [Crossref] [PubMed]
- Hellmann MD, Nathanson T, Rizvi H, et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell 2018;33:843-852.e4. [Crossref] [PubMed]
- Park SE, Park K, Lee E, et al. Clinical implication of tumor mutational burden in patients with HER2-positive refractory metastatic breast cancer. Oncoimmunology 2018;7:e1466768. [Crossref] [PubMed]
- Catenacci DVT, Kang YK, Park H, et al. Margetuximab plus pembrolizumab in patients with previously treated, HER2-positive gastro-oesophageal adenocarcinoma (CP-MGAH22-05): a single-arm, phase 1b-2 trial. Lancet Oncol 2020;21:1066-76. [Crossref] [PubMed]
- Mansfield AS, Aubry MC, Moser JC, et al. Temporal and spatial discordance of programmed cell death-ligand 1 expression and lymphocyte tumor infiltration between paired primary lesions and brain metastases in lung cancer. Ann Oncol 2016;27:1953-8. [Crossref] [PubMed]
- Peng J, Hamanishi J, Matsumura N, et al. Chemotherapy Induces Programmed Cell Death-Ligand 1 Overexpression via the Nuclear Factor-κB to Foster an Immunosuppressive Tumor Microenvironment in Ovarian Cancer. Cancer Res 2015;75:5034-45. [Crossref] [PubMed]
- Bauml J, Seiwert TY, Pfister DG, et al. Pembrolizumab for Platinum- and Cetuximab-Refractory Head and Neck Cancer: Results From a Single-Arm, Phase II Study. J Clin Oncol 2017;35:1542-9. [Crossref] [PubMed]
- Seiwert TY, Burtness B, Mehra R, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol 2016;17:956-65. [Crossref] [PubMed]
- Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362:eaar3593. [Crossref] [PubMed]
- Su S, Zhao J, Xing Y, et al. Immune Checkpoint Inhibition Overcomes ADCP-Induced Immunosuppression by Macrophages. Cell 2018;175:442-457.e23. [Crossref] [PubMed]
- Loi S, Giobbie-Hurder A, Gombos A, et al. Pembrolizumab plus trastuzumab in trastuzumab-resistant, advanced, HER2-positive breast cancer (PANACEA): a single-arm, multicentre, phase 1b-2 trial. Lancet Oncol 2019;20:371-82. [Crossref] [PubMed]
- Emens LA, Esteva FJ, Beresford M, et al. Trastuzumab emtansine plus atezolizumab versus trastuzumab emtansine plus placebo in previously treated, HER2-positive advanced breast cancer (KATE2): a phase 2, multicentre, randomised, double-blind trial. Lancet Oncol 2020;21:1283-95. [Crossref] [PubMed]
- Baselga J, Bradbury I, Eidtmann H, et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): a randomised, open-label, multicentre, phase 3 trial. Lancet 2012;379:633-40. [Crossref] [PubMed]
- Bense RD, Sotiriou C, Piccart-Gebhart MJ, et al. Relevance of Tumor-Infiltrating Immune Cell Composition and Functionality for Disease Outcome in Breast Cancer. J Natl Cancer Inst 2016;109:djw192. [Crossref] [PubMed]


