
LRP6 as a biomarker of poor prognosis of breast cancer
Introduction
Breast cancer is the most common malignancy among women, and it is also the leading cause of cancer-related deaths in women. In 2019, approximately 268,600 cases of invasive breast cancer and 48,100 cases of ductal carcinoma in situ were diagnosed in the United States, and the number of deaths from these types of cancer reached 41,760 (1). Breast cancer has become the main cause of the high mortality of cancer patients because of its high ability of invasion and metastasis. The treatment strategies for breast cancer include local therapy, chemotherapy, targeted therapy, and endocrine therapy. These treatment strategies depend on many factors such as receptor status, tumor type, tumor size, and metastasis. Problems such as triple negative breast cancer, advanced breast cancer, and drug resistance restrict the application of these treatment strategies.
Members of the low-density lipoprotein receptor (LDLR) family include lipoprotein receptor-related protein (LRP)1, LRP5, LRP6, and LRP8. Cancer cells require higher levels of cholesterol than normal cells for their metabolic needs; thus, cancer cells increase their intracellular cholesterol content by enhancing LDLR receptor-mediated endocytosis of serum LDL (2). LDLR has also been reported to have a cancer-promoting effect, facilitating cancer progression through tumor cell migration (3,4). Several studies have shown that the inhibition of LRP1 expression can lead to morphological changes in tumor cells, thus affecting the interaction between cells and cell matrix, specifically by inhibiting focal adhesion kinase (FAK) activation and preventing tumor cell migration (5,6).
The human LRP6 gene is located on chromosome 12p13.2. The full-length 150 kb sequence and its 23 exons are highly conserved in all species. The transmembrane protein LRP6, which acts as a coreceptor, transmits signals through the plasma membrane and activates the Wnt/β-catenin signal pathway (7). The Wnt/β-catenin signaling pathway controls the proliferation, survival, and differentiation of epithelial stem/progenitor cells. Therefore, abnormal activation of this pathway is often observed in cancers of epithelial origin. As an indispensable coreceptor of Wnt, LRP6 is often overexpressed in colorectal cancer, liver cancer and breast cancer, leading to enhancement of the Wnt/β-catenin signaling pathway. In addition, LRP6 hyperphosphorylation has been shown to be increased in KRAS mutant cells and patient-derived colorectal tumors (8).
Based on the results of previous studies, in this study, we hypothesized that the high expression of LRP6 has an adverse effect on the prognosis of breast cancer, by promoting tumor cell invasion and metastasis. In this study, we measured the expression levels of LRP6 in a breast cancer tissue chip, studied the effects of LRP6 on the migration and invasion of the breast cancer cell line MCF-7, and carried out experiments of tumorigenesis in nude mice. These experiments were performed to explore the possibility that LRP6 expression levels in early diagnosis could be markers of poor prognosis of breast cancer. Moreover, this research could provide a theoretical basis for the therapeutic application of LRP6 inhibitors in breast cancer, with inhibition of tumor invasion and metastasis.
We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/gs-21-194).
Methods
Sample and tissue chip and immunohistochemical staining
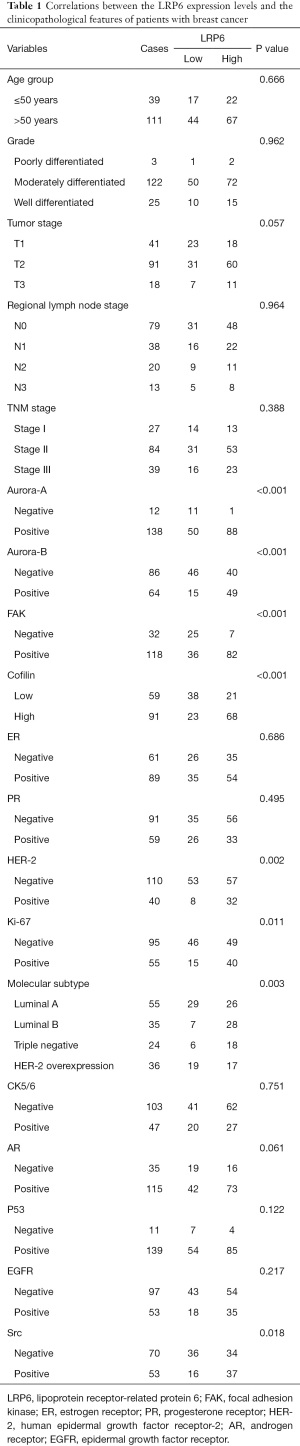
The tissue chips used in this study for immunohistochemical staining were purchased from Shanghai Core Super Co., Ltd. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study design was approved by the Human Research Ethics Committee of Shanghai Outdo Biotech Company (No.: YB M-05-01) and written informed consent had obtained from each patient before their original examination. The clinical characteristics of 150 patients in the tissue chips, such as age, pathological type, pathological grade, tumor invasion, lymph node metastasis, distant metastasis, mitosis, routine immunohistochemistry and survival status, were obtained from medical records (Table 1).
Full table
Immunohistochemical staining was used to detect the expression of LRP6 in breast cancer samples from tissue chips and the mouse tumor tissue. Two experienced pathologists who were blinded to the clinical information of the patients independently evaluated and recorded the immunohistochemical results. The intensity and degree of staining were evaluated in the cytomembrane. LRP6 expression was graded as 0 point (blue), 1 point (yellow), 2 points (brown), and 3 points (brown black), based on tumor cytomembrane staining intensity; grades were assigned based on the percentage of cells that were positive for LRP6 expression as follows: 0 point, 0–5%; 1 point, 6–25%; 2 points, 26–50%; 3 points, 51–75%; 4 points, 76–100%. The scores obtained by optical density and positive percentage analysis (with 0–3 and 4–7 points considered equal to weak and strong expression, respectively) were used to classify LRP6 expression as positive (high staining intensity, >5%), or negative (low staining intensity, <5%) (9). ScanScope chip scanners (Aperio, vista, CA, USA) were used to scan the chips, and Image Scope software (Aperio, vista, CA, USA) was used to collect images of the representative areas. Adobe Illustrator (Adobe) was used for image analysis.
siRNA transfection and construction of lentivirus transfected shRNA-LRP6 low expression cell line
The process of siRNA transfection was as follows. Twenty-four hours before transfection, MCF-7 cells (5×105 cells/hole) were inoculated into a 6-well plate containing 1 mL Opti-MEM (GIBCO, Carlsbad, CA, USA), Subsequently, diluted si-LRP6 and LiPo3000 (Invitrogen, Carlsbad, CA, USA) were mixed with 1:1; further detection was performed after 48 h of culture.
Lentiviral shRNA-LRP6 was purchased from Gemma (Shanghai, China). MCF-7 cells (2–3×105 cells/well) were seeded on a 6-well plate and cultured for 24 h. The viral solution and Polybrene at a final concentration of 5 µg/mL were added into the fresh medium {virus dosage: [number of cells × MOI (multiplicity of infection) value/virus titer] ×103= virus dosage [µL]}. After 12 h, replace the fresh medium.
Colony formation assay
The expression of LRP6 was knocked down by an si-RNA technique in a group of MCF-7 cells obtained from the Chinese Academy of Sciences (ATCC, Beijing, China) (experimental group), while an untreated group of the same series of MCF-7 cells was used as control group. Cells in the logarithmic growth phase were digested with trypsin and seeded on in a petri dish at a density of 500 cells/dish. The petri dish was shaken gently so that the cells were evenly spread; the cells were then grown under standard cell culture conditions at 37 °C in a humidified atmosphere with 5% CO2 using DMEM-H medium (Gibco, Garlsbad, CA, USA) and 10% fetal bovine serum. After 15 days of culture, the cell culture medium was removed, and cells were fixed with pure methanol for 15 minutes, then stained with 5% crystal violet for 20 minutes, after which the number of colonies was counted.
Transwell cell migration assay and cell invasion assay
For the cell migration assay, cells in the logarithmic growth phase were digested with trypsin and diluted to a density of 1×105 cells/mL. An aliquot of 500 µL medium containing 20% fetal bovine serum was added to the lower chamber of a Transwell (Corning Inc., Corning, NY, USA), and a 20 µL aliquot of cell suspension was added to the upper chamber. After 24 hours of culture, 4% paraformaldehyde was used as a fixative for 15 minutes, and the Transwell chamber was stained with 5% crystal violet for 10 min, while the inoculation side of the upper chamber was wiped clean from any remaining cells with cotton swabs. The lower chamber was then photographed under a microscope to observe cell migration. There should have been no fewer than 10 parallel holes in each experimental group. Cells in each visual field were counted and the average value was taken for the final statistical evaluations.
For the cell invasion assays, Matrigel glue was diluted with serum-free medium before cell inoculation. The cells suspension was then slowly added to the Transwell upper chamber containing Matrigel and culture medium and cultured for 2 hours. The following steps of the procedure were the same as those used for the cell migration assays described above.
Wound healing assay
Cells in the logarithmic growth phase were digested with trypsin and inoculated in a pre-labeled 6-well plate. After 24 hours of culture, cells formed an 80% confluent monolayer and were scratched with a 10 µL sterile pipette tip. A culture medium containing 2% fetal bovine serum was used instead of the original medium (to reduce cell proliferation), and photographs were taken at 0, 12 and 24 h.
Tumor model
The nude mice experiments were performed under a project license (No. 2020278) granted by Laboratory Animal Management and Use Committee of Wuhan Seville Biotechnology Co., Ltd., in compliance with guidelines of Hubei province for the care and use of animals. Four-week-old female nude mice purchased from Seville Company (Hubei Province, China) were randomly divided into the lentivirus transfected sh-LRP6 group (experimental group) and sh-NC control group (n=7). Sh-LRP6-MCF-7 and sh-NC-MCF-7 cells were suspended in PBS and mixed with Matrigel in a ratio of 1:1. Estradiol cypionate (2 mg/kg in corn oil, Med Chem Express, Shanghai, China, Cat: HY-B1100) was subcutaneously injected in the chest at the level of the second pair of mammary fat pads on the right side with 1×106 cells. When a visible tumor was formed in the mice, the tumor size was measured once every 3 days, and the tumor volume was calculated according to the formula: volume = (length × width2)/2. The mice were sacrificed 30 days after subcutaneous injection. All the transplanted tumors were removed from the mice and weighed immediately; the expression level of LRP6 in the tumors was then detected by immunohistochemistry. We have previously established the human breast cancer sh-LRP6-MCF-7 (experimental group) and sh-NC-MCF-7 (control group) cell transplantation tumor models in nude mice. Data were collected and analyzed with GraphPad software (GraphPad Software, San Diego, CA, USA) to calculate tumor growth curves.
Statistical analysis
Chi-square test was used to analyze the relationship between LRP6 expression and clinicopathological features of breast cancer. Kaplan-Meier method and logarithmic rank sum test were used for survival analysis in 150 patients with breast cancer, luminal A type breast cancer, luminal B type breast cancer, HER-2 overexpression or triple-negative breast cancer. Cox proportional hazard regression model was used to analyze the potential prognostic factors. All statistical tests were two-tailed tests, where a P<0.05 was considered statistically significant. SPSS Statistics 25.0 software (IBM SPSS, Armonk, NY, USA) was used for all calculations.
Results
Clinicopathological characteristics of patients with breast cancer
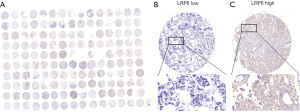
The detailed clinicopathological features of the patients with breast cancer included in this study are shown in Table 1. Of the 150 patients, 111 were over 50 years old. The tumor size and invasion stage (T stage) included T1 stage (n=41), T2 stage (n=91), and T3 stage (n=18); the regional lymph node metastasis stage (N stage) included N0 stage (n=79), N1 stage (n=38), N2 stage (n=20), and N3 stage (n=13); the TNM stage included stage I (n=27), stage II (n=84), and stage III (n=39). Among the molecular subtypes of breast cancer, there were 55 cases of luminal A type, 35 cases of luminal B type, 24 cases of triple-negative breast cancer and 36 cases of HER-2 overexpression. Analysis of routine immunohistochemical indexes revealed that 110 cases were negative for HER-2 and 40 cases were positive; 61 cases were negative for estrogen receptor (ER) and 89 cases were positive; 91 cases were negative for progesterone receptor (PR) and 59 cases were positive. Among the 150 breast cancer tissues by immunohistochemistry staining in tissue microarray (Figure 1A), 61 cases showed low expression of LRP6 (Figure 1B) and 89 cases showed high expression of LRP6 (Figure 1C).
Relationship between LRP6 expression and clinicopathological features of breast cancer
We used the Chi-square test to study the relationship between the expression levels of LRP6 in breast cancer and the clinicopathological features of breast cancer (Table 1). A high expression of LRP6 was significantly related to HER-2 status, Aurora-A, Aurora-B, FAK, Src and Ki67 (P<0.05, Table 1). Among the 110 HER-2 negative patients, 57 patients showed high expression of LRP6, with a proportion of patients displaying high expression of LRP6 equal to 51.82%. Among the 40 HER-2-positive patients, 32 showed high expression of LRP6, with a proportion of patients displaying high expression of LRP6 equal to 80%. The difference between the HER-2 positive and HER-2 negative groups in the percentage of subjects with high LRP6 expression was statistically significant (P=0.002, Table 1). Therefore, the proportion of subjects with high expression of LRP6 among HER-2-positive patients was higher than that in HER-2-negative patients. However, there was no significant correlation between LRP6 expression and tumor size staging, lymph node staging, mitosis, or ER and PR expression (P>0.05, Table 1).
Relationship between LRP6 expression and poor prognosis of breast cancer
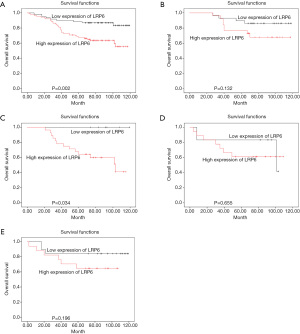
The overall survival rate of 150 patients with breast cancer was analyzed by Kaplan-Meier method and log-rank test. The overall survival rate of tumor patients with high expression of LRP6 was significantly lower than that of patients with low expression of LRP6 (P=0.002, Figure 2A). When stratifying the analysis by the different molecular types of breast cancer, There was no significant difference in overall survival between patients with high expression of LRP6 and those with low expression of LRP6 in patients with breast cancer of luminal A type (P>0.05, Figure 2B); among luminal B type breast cancer patients, the survival rate of patients with high expression of LRP6 was significantly lower than that of patients with low expression of LRP6 (P=0.034, Figure 2C). There was no significant difference in overall survival between patients with high expression of LRP6 and those with low expression of LRP6 in patients with breast cancer of HER-2 overexpression or triple-negative breast cancer (P>0.05, Figure 2D,2E). We used univariate and multivariate Cox risk regression models to assess each risk factor for overall survival (Table 2). Univariate regression analysis showed that positive expression of Aurora-A (HR =2.050, 95% CI: 1.054–3.987, P=0.035), high expression of LRP6 (HR =1.647, 95% CI: 1.108–2.448, P=0.014), and high expression of Cofilin (HR =1.510, 95% CI: 1.017–2.240, P=0.041) were negative prognostic factors for breast cancer. In multivariate regression analysis, LRP6 was an independent risk factor for breast cancer, and the risk of death in the LRP6 high expression group was 1.653 times higher than that in the low expression group (95% CI: 1.039–2.631, P= 0.034).
Full table
Effect of si-RNA knockdown of LRP6 on breast cancer cells cell clone formation
We used RT-qPCR to detect the expression of LRP6 in MCF-7 and MCF-10A cell lines. The results showed that the expression of LRP6 in breast cancer cells was significantly higher than that in breast epithelial cells (Figure 3A). We used siRNA to knockdown the expression of LRP6 in MCF-7 cell lines (Figure 3B).
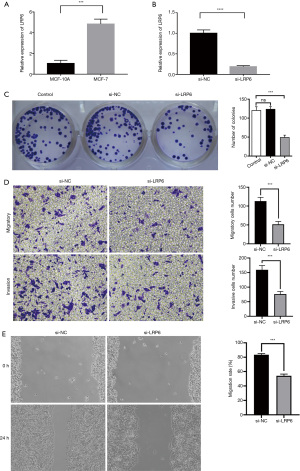
In order to further investigate the effect of LRP6 on the proliferation of breast cancer cells, we carried out a cell clone formation assay. In the MCF-7 cell line, LRP6 knockdown significantly reduced the cell clone formation rate as compared to that in the control group (P<0.05, Figure 3C).
Effects of si-RNA knockdown of LRP6 on breast cancer cell migration, invasion, and wound healing
The results of Transwell assay showed that the migration and invasion of cells were significantly inhibited in the si-RNA knockdown LRP6 group as compared to that in the control group (P<0.05, Figure 3D). In the scratch experiment, the wound healing ability of the experimental group was significantly weaker than that of the control group (P<0.05, Figure 3E).
Study on the relationship between LRP6 expression and transplanted tumors in nude mice
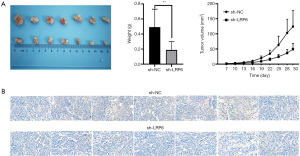
We studied the effect of LRP6 on tumor formation in nude mice by constructing human breast cancer sh-LRP6-MCF-7 (experimental group) and sh-NC-MCF-7 (control group) cell transplantation tumor models, we observed that the tumor growth rate of the experimental group was significantly lower than that of the control group; the size and weight of the 7 tumors that developed in nude mice in the experimental group were significantly smaller than those of the control group. There was a significant difference in tumor growth volume and weight between the two groups (P<0.05, Figure 4A). We also used the immunohistochemistry staining to detect the expression of LRP6 in the transplanted tumors of the experimental group and the control group (Figure 4B).
Discussion
In recent years, individualized diagnosis and treatment and molecular targeted therapy for breast cancer patients have become topics of intense research. How to improve the overall survival rate and long-term prognosis of breast cancer patients has increasingly become the focus of clinicians. The most common subtype of breast cancer is ER positive luminal subtype, which accounts for 80% of cases of breast cancer. Current treatment for luminal subtype includes surgical removal, chemotherapy, radiotherapy and endocrine therapy. Despite the promising initial response to treatment, the recurrence, metastasis, and drug resistance of tumor are still the main causes of death among these patients (10). Therefore, there is an urgent need to develop new therapeutic targets for these patients.
The immunohistochemical staining performed in this study showed that in the tissue microarray of 150 cases of breast cancer, the proportion of patients with a high expression level of LRP6 was 59.3%. Previous studies have found that the percentage of patients showing high expression levels of LRP6 in colorectal cancer, hepatocellular carcinoma and breast cancer are similar to that found in our study (8).
In this study, we found that age, tumor size stage, lymph node stage and mitosis were not significantly correlated with the expression of LRP6, suggesting that the expression of LRP6 is not influenced by these pathological factors in promoting the occurrence and development of breast cancer. Through univariate and multivariate regression analyses, we found that the high expression of LRP6 was an independent risk factor for breast cancer, and that it was positively correlated with mortality and poor prognosis of breast cancer. In agreement with what we observed in this study, previous studies revealed that high expression of LRP6 is associated with a malignant phenotype, metastatic potential, and poor prognosis also for epithelial tumors, such as hepatocellular carcinoma, oral squamous cell carcinoma, and colorectal cancer (11-15). Previous studies found that the expression level of LRP6 in ER, PR, and HER-2 negative breast tumors is higher than that in ER, PR, and HER-2 positive breast tumors (16). In this study, we observed, no significant correlation between the expression of ER and PR and the expression of LRP6. The percentage of cases with high expression of LRP6 was higher among HER-2 positive than that among HER-2 negative patients, suggesting that LRP6 can still contribute to promotion, and development of breast cancer when ER, PR and HER-2 are positive.
Survival analysis showed that a high expression of LRP6 in breast cancer tissue was negatively correlated with the overall survival rate. When considering the molecular type of breast cancer, the expression of LRP6 in patients with luminal B type breast cancer was negatively correlated with the overall survival rate; however, in other molecular types, especially triple-negative breast cancer, there was no significant difference in overall survival rate between patients with high expression of LRP6 and those with low expression of LRP6. In our study, the results show that LRP6 may be an important factor affecting the occurrence and progression of breast cancer, and that the detection of LRP6 expression in breast cancer tissue samples is helpful for early diagnosis and prognosis. Similar conclusions were previously reported in the studies of colorectal cancer (17,18).
The results of this study demonstrated that after knocking down LRP6, the clone formation rate, cell migration ability, invasion ability and wound healing ability of the breast cancer MCF-7 cell line was significantly lower than those of the control group. Similar experimental results were also found in bladder cancer cell line T24 (19). The results reported in the current literature suggest that the expression of LRP6 is up-regulated in human triple-negative breast cancer patients and human TNBC cell lines; conversely, the knockdown of LRP6 expression and treatment with recombinant MESD protein (a specific inhibitor of LRP6) significantly reduced the migration and invasion of TNBC MDA-MB-231 and BT549 cells (16). These previous results are similar to our results, but our study suggests that the high expression of LRP6 can still promote the proliferation, migration, and invasion of breast cancer cells when ER is positive. In our tumorigenic model in nude mice, inhibition of LRP6 expression led to inhibition of tumor growth, suggesting that LRP6 plays a key role in tumor proliferation. Ettenberg et al. reported that specific LRP6 antibodies or the LRP6 inhibitors MESD inhibit the growth of WNT1-or WNT3a-driven xenografts (20). These results suggest that LRP6 may be a new target for the treatment of breast cancer.
Previous studies have shown that natural compounds isolated from the plants, such as prodigiosin, silibinin, rottlerin, salinomycin and gigantol, inhibit the activity of Wnt/β-catenin in TNBC cells by inhibiting the expression and phosphorylation of LRP6 (21-25). In addition, studies suggest that niclosamide induces LRP6 degradation in breast cancer cell lines, resulting in an increase in tumor cell apoptosis and a decrease in tumor cell proliferation (26).
Conclusions
Therefore, LRP6 can become a molecular targeting site for breast cancer therapy, and LRP6 inhibitors can be used in the treatment of breast cancer. Exploring the basic theory of the application of LRP6 inhibitors in breast cancer will be helpful towards the achievement of personalized targeted therapy for breast cancer patients.
Acknowledgments
We would like to thank Editage (
Funding: This work was supported by National Nature Science Foundation of China (grant number: 81702650).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/gs-21-194
Data Sharing Statement: Available at https://dx.doi.org/10.21037/gs-21-194
Peer Review File: Available at https://dx.doi.org/10.21037/gs-21-194
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/gs-21-194). All authors report that this work was supported by National Nature Science Foundation of China (grant number: 81702650).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics committee of Shanghai Outdo Biotech Company (No.: YB M-05-01) and informed consent was taken from all the patients. Experiments were performed under a project license (No.: 2020278) granted by Laboratory Animal Management and Use Committee of Wuhan Seville Biotechnology Co., Ltd., in compliance with guidelines of Hubei province for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- DeSantis CE, Ma J, Gaudet MM, et al. Breast cancer statistics, 2019. CA Cancer J Clin 2019;69:438-51. [Crossref] [PubMed]
- Zhou T, Zhan J, Fang W, et al. Serum low-density lipoprotein and low-density lipoprotein expression level at diagnosis are favorable prognostic factors in patients with small-cell lung cancer (SCLC). BMC Cancer 2017;17:269. [Crossref] [PubMed]
- Jiang L, Jiang S, Lin Y, et al. Combination of body mass index and oxidized low density lipoprotein receptor 1 in prognosis prediction of patients with squamous non-small cell lung cancer. Oncotarget 2015;6:22072-80. [Crossref] [PubMed]
- Migita T, Ruiz S, Fornari A, et al. Fatty acid synthase: a metabolic enzyme and candidate oncogene in prostate cancer. J Natl Cancer Inst 2009;101:519-32. [Crossref] [PubMed]
- Appert-Collin A, Bennasroune A, Jeannesson P, et al. Role of LRP-1 in cancer cell migration in 3-dimensional collagen matrix. Cell Adh Migr 2017;11:316-26. [Crossref] [PubMed]
- Dedieu S, Langlois B, Devy J, et al. LRP-1 silencing prevents malignant cell invasion despite increased pericellular proteolytic activities. Mol Cell Biol 2008;28:2980-95. [Crossref] [PubMed]
- Roslan Z, Muhamad M, Selvaratnam L, et al. The Roles of Low-Density Lipoprotein Receptor-Related Proteins 5, 6, and 8 in Cancer: A Review. J Oncol 2019;2019:4536302 [Crossref] [PubMed]
- Raisch J, Côté-Biron A, Rivard N. A Role for the WNT Co-Receptor LRP6 in Pathogenesis and Therapy of Epithelial Cancers. Cancers (Basel) 2019;11:1162. [Crossref] [PubMed]
- Maimaiti Y, Liu Z, Tan J, et al. Dephosphorylated cofilin expression is associated with poor prognosis in cases of human breast cancer: a tissue microarray analysis. Onco Targets Ther 2016;9:6461-6. [Crossref] [PubMed]
- Kumar S, Srivastav RK, Wilkes DW, et al. Estrogen-dependent DLL1-mediated Notch signaling promotes luminal breast cancer. Oncogene 2019;38:2092-107. [Crossref] [PubMed]
- González-Sancho JM, Aguilera O, García JM, et al. The Wnt antagonist DICKKOPF-1 gene is a downstream target of beta-catenin/TCF and is downregulated in human colon cancer. Oncogene 2005;24:1098-103. [Crossref] [PubMed]
- Jia Q, Bu Y, Wang Z, et al. Maintenance of stemness is associated with the interation of LRP6 and heparin-binding protein CCN2 autocrined by hepatocellular carcinoma. J Exp Clin Cancer Res 2017;36:117. [Crossref] [PubMed]
- Liu Z, Sun B, Qi L, et al. Dickkopf-1 expression is down-regulated during the colorectal adenoma-carcinoma sequence and correlates with reduced microvessel density and VEGF expression. Histopathology 2015;67:158-66. [Crossref] [PubMed]
- Qi L, Sun B, Liu Z, et al. Dickkopf-1 inhibits epithelial-mesenchymal transition of colon cancer cells and contributes to colon cancer suppression. Cancer Sci 2012;103:828-35. [Crossref] [PubMed]
- Yuan Y, Xie X, Jiang Y, et al. LRP6 is identified as a potential prognostic marker for oral squamous cell carcinoma via MALDI-IMS. Cell Death Dis 2017;8:e3035 [Crossref] [PubMed]
- Ma J, Lu W, Chen D, et al. Role of Wnt Co-Receptor LRP6 in Triple Negative Breast Cancer Cell Migration and Invasion. J Cell Biochem 2017;118:2968-76. [Crossref] [PubMed]
- Lemieux E, Cagnol S, Beaudry K, et al. Oncogenic KRAS signalling promotes the Wnt/β-catenin pathway through LRP6 in colorectal cancer. Oncogene 2015;34:4914-27. [Crossref] [PubMed]
- Yao Q, An Y, Hou W, et al. LRP6 promotes invasion and metastasis of colorectal cancer through cytoskeleton dynamics. Oncotarget 2017;8:109632-45. [Crossref] [PubMed]
- Jiang F, Qi W, Wang Y, et al. lncRNA PEG10 promotes cell survival, invasion and migration by sponging miR-134 in human bladder cancer. Biomed Pharmacother 2019;114:108814 [Crossref] [PubMed]
- Ettenberg SA, Charlat O, Daley MP, et al. Inhibition of tumorigenesis driven by different Wnt proteins requires blockade of distinct ligand-binding regions by LRP6 antibodies. Proc Natl Acad Sci U S A 2010;107:15473-8. [Crossref] [PubMed]
- Lu W, Li Y. Salinomycin suppresses LRP6 expression and inhibits both Wnt/β-catenin and mTORC1 signaling in breast and prostate cancer cells. J Cell Biochem 2014;115:1799-807. [Crossref] [PubMed]
- Lu W, Lin C, King TD, et al. Silibinin inhibits Wnt/β-catenin signaling by suppressing Wnt co-receptor LRP6 expression in human prostate and breast cancer cells. Cell Signal 2012;24:2291-6. [Crossref] [PubMed]
- Lu W, Lin C, Li Y. Rottlerin induces Wnt co-receptor LRP6 degradation and suppresses both Wnt/β-catenin and mTORC1 signaling in prostate and breast cancer cells. Cell Signal 2014;26:1303-9. [Crossref] [PubMed]
- Wang Z, Li B, Zhou L, et al. Prodigiosin inhibits Wnt/β-catenin signaling and exerts anticancer activity in breast cancer cells. Proc Natl Acad Sci U S A 2016;113:13150-5. [Crossref] [PubMed]
- Yu S, Wang Z, Su Z, et al. Gigantol inhibits Wnt/β-catenin signaling and exhibits anticancer activity in breast cancer cells. BMC Complement Altern Med 2018;18:59. [Crossref] [PubMed]
- Lu W, Lin C, Roberts MJ, et al. Niclosamide suppresses cancer cell growth by inducing Wnt co-receptor LRP6 degradation and inhibiting the Wnt/β-catenin pathway. PLoS One 2011;6:e29290 [Crossref] [PubMed]


