
A plasma-derived extracellular vesicle mRNA classifier for the detection of breast cancer
Introduction
According to the latest global cancer burden data released by the World Health Organization (WHO) in 2020, the number of new cases of breast cancer (BC) has increased rapidly, replacing lung cancer as the most common cancer, and comprising 11.7% of all cancer cases. Among women, BC is the most frequently diagnosed cancer, accounting for approximately 25% of cancer cases and is the main cause of cancer death (1). Similar to other types of cancer, early diagnosis can reduce the incidence and mortality rates of BC, and is the key to improving the survival rate. Therefore, early diagnosis of BC has become an important aspect in the treatment of BC.
In various platforms, imaging techniques are major diagnostic methods and include ultrasound scanning, mammography, magnetic resonance imaging (MRI), and positron-emission tomography (PET), which can provide valuable data for the treatment of BC patients. However, some of these techniques are expensive, lack sensitivity for detecting early BC, and/or expose the patient to radiation, which adds an extra risk factor. In addition, some of the most studied and established BC biomarkers, such as CEA, CA125, and CA15-3, have been used to detect and monitor the diagnosis of BC in patients; however, low sensitivity limits their applications in early BC detection (2). Moreover, the currently available diagnostic technologies for use with BC have certain limitations, and the probability of false-positive results is 20–50% (3). Therefore, there is an urgent need for non-invasive methods that can more sensitively and specifically identify BC in the early stage.
Extracellular vesicles (EVs) produced by cells are mainly divided into three types according to their size and biogenesis: exosomes (40–200 nm), microvesicles (200–1,000 nm) and apoptotic bodies (500–2,000 nm) (4). EVs play an important role in breast physiology and pathophysiology including carcinogenesis, tumor progression, molding the tumor microenvironment, and immune modulation. Exosomes, as a class of EVs, are nano-vesicles related to the physiologic regulation of breast development (such as lactation), but they are also significant mediators of breast tumors, endowing them with great potential as new molecular markers for the diagnosis and prognosis of BC (5). Exosomes are considered to be important mediators of cell-to-cell communication, through the horizontal transfer of bioactive substances, such as proteins (6), messenger RNA (mRNA) and non-coding RNA (7). Considerable evidence has suggested that mRNAs enriched in serum exosomes can be effectively internalized recipient cells (8,9). Therefore, the mRNA in exosomes is a potential biomarker for use in liquid biopsy, and the use of exosomal profiling without tissue has obvious prospects for the early diagnosis of diseases. Up to now, there is no research on developing BC diagnosis model by detecting multiple plasma-derived EV mRNAs.
In this study, we exploited a non-invasive method for the diagnosis of BC. First, we employed next-generation sequencing to determine the RNA profiles of plasma EVs derived from 14 patients with BC and 6 patients with benign breast lesions serving as controls. Next, we compared these RNA profiles to identify the mRNAs of plasma EVs at significantly different levels. Finally, we developed predictive classifiers with multiple machine learning models to discriminate patients with BC from individuals without cancer. In summary, our study may provide some new suggestions for the diagnosis of BC.
We present the following article in accordance with the STARD reporting checklist (available at https://dx.doi.org/10.21037/gs-21-275).
Methods
Sample characteristics
A total of 259 plasma samples were derived from 3 groups of participants enrolled at The First People’s Hospital of Foshan, including 144 with BC, 72 benign with breast lesions, and 43 healthy women. All the plasma samples of cancer patients were prospectively collected during the discovery stage and before cancer therapy was initiated. The plasma samples of the discovery cohort were prospectively collected between January 2018 and July 2018. The samples comprising the training and validation cohorts were prospectively collected between August 2018 and May 2019. With the informed consent of all participants, all plasma samples were collected for research according to the protocols approved by the Ethics Committee of the First People’s Hospital of Foshan (ID: L[2021]-7). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Details regarding the clinical information of the participants involved in this study are listed in Table S1.
RNA isolation from Plasma EVs
Total RNA was isolated using an exoRNeasy Serum/Plasma Maxi Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. We purified exosomes and other EVs and isolated total versicular RNA, including mRNA, non-coding RNA, miRNA and other small RNAs, from 2–4 mL serum. In brief, the plasma was prefiltered, then was mix with 2× binding buffer. The mixture is added to the exoEasy membrane affinity to bind the EVs to the membrane. After centrifugation, the wash buffer was added to wash off non-specific material in the column. After enriching EVs, QIAZOL was added to the column to lyse the vesicles and chloroform was added to the lysate collected after centrifugation. The aqueous phases os recovered and mixed with ethanol, and the sample-ethanol mixture is added to the RNeasy MinElute spin column and centrifuged. Wash the column with buffer RWT, then wash twice with buffer RPE. And finally elute RNA in water.
Library preparation and next-generation sequencing
A high-throughput sequencing service, transcriptome high-throughput sequencing, and subsequent bioinformatics analysis were provided by Cloud-Seq Biotech (Shanghai, China). Briefly, total RNA was used to remove ribosomal RNA (rRNA) prior to the construction of the RNA-seq libraries with Ribo-Zero rRNA Removal Kits (Illumina, San Diego, CA, USA) following the manufacturer’s instructions. The RNA libraries were prepared using a TruSeq Stranded Total RNA Library Prep Kit (Illumina) with rRNA-depleted RNAs following the manufacturer’s instructions. A BioAnalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA) was used to quantify and control the quality of the libraries. According to the manufacturer’s instructions, 10 pM libraries were denatured as single-stranded DNA molecules, captured in Illumina flow cells, and amplified into clusters in situ. Finally, each nucleic acid sequence was sequenced for 150 cycles on an Illumina HiSeq Sequencer.
Data analysis of RNA-sequencing
Paired-end reads were obtained from an Illumina HiSeq 4000 sequencer, and quality control was performed by Q30. Low-quality reads were filtered by Cutadapt software (v1.9.3) after 3’ adaptor trimming (9). The high-quality trimmed reads were used to analyze mRNAs and were aligned to the human reference genome (UCSC hg19) using HISAT2 software v2.0.4 (http://www.ccb.jhu.edu/software/hisat/index.shtml) (10). The FPKM was obtained by using Cuffdiff software (v2.2.1, in the Cufflinks package) (10) and used for the mRNA expression profiles as guided by the Ensembl gtf gene annotation file. Then, fold change and P value were determined based on the FPKM to identify the differentially expressed mRNAs. We performed GO and pathway enrichment analyses with the clusterProfiler package on the mRNAs expressed at differential levels.
Quantitative real-time polymerase chain reaction
Complementary DNA was synthesized using a PrimeScript RT reagent Kit (Takara, Bio Inc., Kusatsu, Shiga, Japan) after RNA isolation. mRNA quantification was obtained with qPCR using SYBR Premix Ex Taq (Takara, Bio Inc.) and an ABI7500 Real-Time PCR system (Applied Biosystems, Forster City, CA, USA) with the primers and complementary DNA (cDNA) template recommended by the manufacturer. The primers are listed in Table S2.
Classifier construction
To develop classifiers for BC diagnosis, 13 candidate mRNAs with significantly elevated levels in BC patients were selected and assessed using quantitative real-time polymerase chain reaction (qRT-PCR). A total of 259 participants were divided into training and validation cohorts at a ratio of 7:3. The training cohort contained 182 participants, including 101 patients with BC and 81 without cancer serving as controls (51 individuals with benign breast lesions and 30 healthy women). The validation cohort contained 77 participants, including 43 patients with BC and 34 without cancer serving as controls (21 individuals with benign breast lesions and 13 healthy women). To construct predictive classifiers that can be used to differentiate individuals with BC from those serving as controls, we used 3 regression models: support vector machine (SVM), logistic regression (LR), and linear discriminate analysis (LDA). As numerous studies have reported that discrete data may improve classifier performance (11), continuous variables were discretized before classifier construction according to the optimal cut-off point, which was selected according to the maximum value of (sensitivity + specificity)/2 in the training cohort. Then, the continuous value was set to 1 when it was higher than the corresponding optimal cut-off for each subject; otherwise, it was set to 0 (Table S3). Using the stepwise method, the optimal classifier with the largest AUC was selected. The leave-1-out cross-validation (LOOCV) method was used to estimate the robustness and prediction error of the selected classifiers. In the validation cohort, the relative levels of the mRNAs in the selected classifiers were determined, and the predictive efficacy of the classifiers was evaluated in the test cohort.
Statistical analysis
We used R software (version 3.4; https://www.R-project.org/) for statistical analyses. Mann-Whitney U test or Kruskal-Wallis test was used to compare the differences in patient characteristics in 2 or more groups, respectively, and Pearson chi-square was used to compare sample compositions. The diagnostic accuracy of the candidate mRNAs or their combinations was evaluated by receiver operating characteristic (ROC) curve analysis with the area under the ROC curve (AUC). A P value <0.05 was considered significant.
Results
Significantly differential mRNAs identification
To identify different levels of plasma EV mRNAs between patients with BC and those without cancer, we first generated the mRNA profiles of the exosomes obtained from the plasma of 14 participants with BC and 6 with benign lesions during the discovery stage. We found that the levels of 1,156 mRNAs were significantly different, including 183 mRNAs at higher levels and 973 mRNAs at lower levels (Figure 1A, Table S4, |log2-fold change| >2, P value <0.05). By implementing unsupervised clustering analysis, we found distinct patterns between the participants with and without cancer, and the samples in each group clustered together in separate groups (Figure 1B), which indicated the potential of using mRNAs from EVs for screening BC.
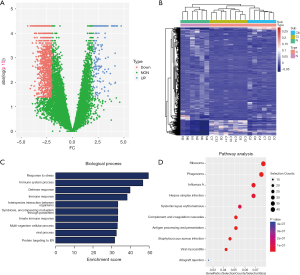
Gene function and pathway enrichment analyses
To reveal the potential roles of mRNAs as indicated by their different levels in cases of tumorigenesis, we performed a gene function enrichment analysis. The results indicated that the terms associated with these mRNAs were enriched in multiple processes, such as multiple biological pathways and processes related to immunity (Figure 1C). For example, HCK participates in a variety of biological processes by mediating immune response regulation signaling pathways and multiple signaling pathways [including epithelial to mesenchymal transition (EMT), the PI3K/AKT signaling pathway, and focal adhesion] in BC (12). In addition, AXL is involved in a variety of processes that promote tumor formation and metastasis, and is considered an effective biomarker for BC. Especially in triple negative (TN)BC tumors, AXL is considered a key factor in promoting EMT associated with mesenchymal and invasive phenotypes (13). In addition, we performed a pathway enrichment analysis and found that some of the mRNAs in BC are closely related to ribosomes (Figure 1D). These mRNAs may play important roles in the translation process.
Classifier construction
For clinical practice and detection convenience, we focused on the mRNAs of plasma EVs with elevated levels. According to fold change, the false discovery rate (FDR) value and mean levels, we selected 12 mRNAs for further qRT-PCR identification and predictive classifier construction. To develop predictive classifiers for BC diagnosis, the samples were split into training and validation cohorts. Using qRT-PCR, we assessed the relative levels of 12 mRNAs in the training group that were at higher levels in the 182 plasma samples obtained from 101 BC participants and 81 patients without cancer serving as controls (51 participants with benign breast lesions and 30 healthy women). We used 3 regression models, SVM, LDA, and LR, to construct mRNA classifiers that could be used to differentiate individuals with BC from those without cancer serving as controls. The AUC and accuracy, sensitivity, and specificity of the classifiers were cross-validated with the LOOCV method (Figure 2). Among all combinations assessed with the 3 different regression models, an 8-mRNA combination, named EXOBmRNA, achieved high performance [AUC =0.718, 95% confidence interval (CI): 0.652 to 0.784, and accuracy =71.9%] in the training cohort after LOOCV was performed, showing the largest AUC with the SVM model (Figure 2A). The mRNAs in EXOBmRNA were HLA-DRB1, HAVCR1, ENPEP, TIMP1, CD36, MARCKS, DAB2, and CXCL14. Then, the performance of EXOBmRNA was subsequently studied and verified in the validation cohort. In the validation cohort, the AUC for the EXOBmRNA group was 0.737 (95% CI: 0.636 to 0.837, Figure 2E,F). In contrast with that of the training cohort, the AUC of the validation cohort was similar to that of the training cohort (P=0.764).
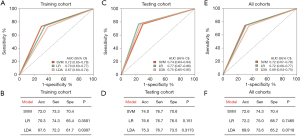
Discussion
Our study found 1,156 mRNAs at different levels between 14 participants with BC and 6 participants with benign breast lesions by comparing the mRNA profiles using next-generation sequencing. In addition, gene function and pathway analyses showed that the genes expressed at differential levels were related to cancer. Finally, we developed predictive classifiers to assess their potential for BC diagnosis, and we found that the AUC of the EXOBmRNA group was 0.724 (0.669 to 0.779).
Our method is non-invasive, and detection is not confounded by the heterogeneity of a tumor. At the same time, it does not expose patients to the effects of radiation and shows high sensitivity and specificity, highlighting great potential for use in the early screening of BC. However, this method has certain limitations. The cost of extracting exosomes is still very high, kits are expensive, and an ultracentrifuge is often lacking in the clinic; therefore, this method is still difficult to use extensively in the clinic. In addition, the classifier model needs to be further optimized, and the biological function of each mRNA and the relationship between them need to be further studied. With optimization and further characterization, the possibility that this method will be used in clinical applications will be increased.
We searched the literature for the genes that we identified and discovered that many of the mRNAs and their translated proteins are closely related to BC (14). For instance, DAB2 can be used as a novel marker and therapeutic target for BC (15), MARCKS is closely related to poor prognosis of BC, and knockdown of ENPEP can inhibit the invasion of BC (16). In addition, there have been many research reports on these genes in other tumors. First, they can be used as tumor risk genes or prognostic genes, and their expression levels in tumor patients are significantly increased. For example, HLA-DRB1 is considered a risk gene for cervical cancer (17), and TIMP1 may be related to the metastasis of uveal melanoma (UM), possibly playing an important role in identifying patients with high metastatic risk and predicting the prognosis of UM (18). Second, these genes are important for the occurrence and development of tumors. For instance, CD36 upregulates DEK transcription and promotes cell migration and invasion and EMT in gastric cancer (19). Overexpression of HAVCR1 can reduce cell adhesion and invasion in colorectal cancer (8). Finally, these genes can play important roles in tumor immunity and the tumor microenvironment and are potential targets for future immunotherapy. For instance, the chemokine CXCL14 is often dysregulated in several types of cancer, and its destructive effect limits key antitumor immune regulator function and is closely related to poor patient prognosis.
Therefore, we expect that the biological functions and characteristics of these mRNAs can be combined to construct different classifiers for use in early tumor diagnosis or subtype classification of tumors.
Acknowledgments
Funding: The work was supported by the Medical Scientific Research Foundation of Guangdong Province of China (B2017006); Special fund of Foshan Summit plan (2020G010); and Foshan City medical science and technology project (2020001005030).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://dx.doi.org/10.21037/gs-21-275
Data Sharing Statement: Available at https://dx.doi.org/10.21037/gs-21-275
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/gs-21-275). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. With the informed consent of all participants, all plasma samples were collected for research according to the protocols approved by the Ethics Committee of the First People’s Hospital of Foshan (ID: L[2021]-7). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Jafari SH, Saadatpour Z, Salmaninejad A, et al. Breast cancer diagnosis: Imaging techniques and biochemical markers. J Cell Physiol 2018;233:5200-13. [Crossref] [PubMed]
- Lindenberg MA, Miquel-Cases A, Retèl VP, et al. Imaging performance in guiding response to neoadjuvant therapy according to breast cancer subtypes: A systematic literature review. Crit Rev Oncol Hematol 2017;112:198-207. [Crossref] [PubMed]
- Kim KM, Abdelmohsen K, Mustapic M, et al. RNA in extracellular vesicles. Wiley Interdiscip Rev RNA 2017;8: [Crossref] [PubMed]
- Zha QB, Yao YF, Ren ZJ, et al. Extracellular vesicles: An overview of biogenesis, function, and role in breast cancer. Tumour Biol 2017;39:1010428317691182 [Crossref] [PubMed]
- Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002;2:569-79. [Crossref] [PubMed]
- Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [Crossref] [PubMed]
- Wang Y, Martin TA, Jiang WG. HAVcR-1 expression in human colorectal cancer and its effects on colorectal cancer cells in vitro. Anticancer Res 2013;33:207-14. [PubMed]
- Mitsuhashi M, Taub DD, Kapogiannis D, et al. Aging enhances release of exosomal cytokine mRNAs by Aβ1-42-stimulated macrophages. FASEB J 2013;27:5141-50. [Crossref] [PubMed]
- Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012;7:562-78. [Crossref] [PubMed]
- Lin XJ, Chong Y, Guo ZW, et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol 2015;16:804-15. [Crossref] [PubMed]
- Zhu X, Zhang Y, Bai Y, et al. HCK can serve as novel prognostic biomarker and therapeutic target for Breast Cancer patients. Int J Med Sci 2020;17:2773-89. [Crossref] [PubMed]
- Falcone I, Conciatori F, Bazzichetto C, et al. AXL Receptor in Breast Cancer: Molecular Involvement and Therapeutic Limitations. Int J Mol Sci 2020;21:8419. [Crossref] [PubMed]
- Tian X, Zhang Z. miR-191/DAB2 axis regulates the tumorigenicity of estrogen receptor-positive breast cancer. IUBMB Life 2018;70:71-80. [Crossref] [PubMed]
- Manai M, Abdeljaoued S, Goucha A, et al. MARCKS protein overexpression is associated with poor prognosis in male breast cancer. Cancer Biomark 2019;26:513-22. [Crossref] [PubMed]
- Feliciano A, Castellvi J, Artero-Castro A, et al. miR-125b acts as a tumor suppressor in breast tumorigenesis via its novel direct targets ENPEP, CK2-α, CCNJ, and MEGF9. PLoS One 2013;8:e76247 [Crossref] [PubMed]
- Kamiza AB, Kamiza S, Mathew CG. HLA-DRB1 alleles and cervical cancer: A meta-analysis of 36 case-control studies. Cancer Epidemiol 2020;67:101748 [Crossref] [PubMed]
- Wang P, Yang X, Zhou N, et al. Identifying a Potential Key Gene, TIMP1, Associated with Liver Metastases of Uveal Melanoma by Weight Gene Co-Expression Network Analysis. Onco Targets Ther 2020;13:11923-34. [Crossref] [PubMed]
- Wang J, Wen T, Li Z, et al. CD36 upregulates DEK transcription and promotes cell migration and invasion via GSK-3β/β-catenin-mediated epithelial-to-mesenchymal transition in gastric cancer. Aging (Albany NY) 2020;13:1883-97. [Crossref] [PubMed]


