
Predictive value of lncRNA LOC100505851 in breast cancer in the neoadjuvant setting
Introduction
Breast cancer is the leading cause of all tumor-associated deaths in women worldwide as well as in China nationwide. Around 5–30% of the patients came with locally advanced disease (1,2), which is not immediately operable and calls for presurgical systematic treatment known as neoadjuvant therapy (NAT). In addition to increasing the chances of effective surgery, NAT provides valuable information about the patient’s response to the current therapy, which could guide further clinical decisions. The response to NAT is measured as pathological complete response (pCR), defined as no residual tumor cells in the surgical specimen by pathological assessment (3). Achievement of pCR indicates sensitivity to the current treatment and a better prognosis, while other patients might need to switch to an alternative treatment after surgery (4,5). pCR is also considered a surrogate for survival and accelerated drug approval (6). Thus, it is important to identify biomarkers to predict pCR early to improve the effect and efficiency of NAT.
Long noncoding RNAs (lncRNAs) are a type of non-protein-coding RNA transcript with a length of more than 200 bp. Although lncRNAs do not encode proteins directly, they can still participate and function in various kinds of biological pathways, such as genetic imprinting, epigenetic modification, chromatin conformation, cell cycle regulation, stem cell differentiation, and the regulation of enzyme activity in a cis- or trans-manner (7,8). In recent years, studies have found that lncRNAs also play a crucial role in oncogenesis (9). The expression of lncRNAs in tumors is tissue-specific, and they can act as oncogenes or tumor suppressor genes at the transcriptional and/or posttranscriptional levels, thereby affecting tumor cell proliferation, apoptosis, angiogenesis, invasion and metastasis (9,10). Therefore, lncRNAs can be used as important molecular biomarkers for tumor diagnosis and prediction (11).
LncRNA LOC100505851 is located on chr19p12. It was first identified by the Mammalian Gene Collection Program Team in 2002 when they analyzed more than 15,000 full-length human and mouse cDNA sequences; just one year later, Toshio Ota and his colleagues reported the existence of this unique lncRNA after they had sequenced and characterized 21,243 full-length human cDNAs (12,13). LOC100505851 is the antisense transcript of zinc finger protein 726 (ZNF 726). ZNFs belong to one of the largest known transcript families and are characterized by their unique zinc finger structure (14). ZNF 726 was postulated to be a tumor suppressor gene in colorectal cancer (CRC) due to its downregulation in tumor tissue and its positive correlation with overall survival (OS) (15).
Currently, the function of lncRNA LOC100505851 in breast cancer is unknown. To explore and clarify the expression of this novel lncRNA and its clinical significance, we conducted this retrospective study using data from prospective neoadjuvant clinical trials in breast cancer and preliminarily explored its underlying mechanisms.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/gs-21-3).
Methods
Patients and study design
A total of 209 patients with adequate and qualified tissue samples for the detection of lncRNA LOC100505851 expression were enrolled. Of which 106 patients receiving surgery (without NAT) at Renji Hospital from March 1, 2014 to December 31, 2015, provided with both adjacent and cancer tissue, were enrolled for correlational analysis. Another 103 patients enrolled between June 1, 2014 and October 31, 2018 in two prospective neoadjuvant clinical trials [registered in the ClinicalTrials. gov website as SHPD001 (ClinicalTrials.gov identifier: NCT02199418) and SHPD002 (ClinicalTrials.gov identifier: NCT02221999)] were collected for both correlation and pCR analysis, providing only cancer tissue. Details of the neoadjuvant treatment protocols have been reported previously (16). The study protocols were approved by the ethics committee of Renji Hospital (IRB approval No. 2014-14K and 2017-088). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). And written informed consent of donating biospecimen for scientific researchers were obtained from all the study participants.
Cell culture and subcellular localization
Human breast cancer cell line ZR7530 was obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cell lines were maintained in McCoy’s 5A and Leibovitz’s L-15 medium (BasalMedia, Shanghai, China). All media were supplemented with 10% fetal bovine serum (Gibco, Waltham, MA, USA) and 1% penicillin-streptomycin (BasalMedia, Shanghai, China). Nuclear and cytoplasmic RNA extractions were carried out in the ZR7530 cell lines. The cytoplasm expressed 18s rRNA and the nuclear expressed U6 were used as controls. The experimental procedure was carried out according to the relevant instructions of the NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Thermo Scientific, USA).
RNA extraction and PCR
Total RNA was extracted from tissues using TRIzol reagent (Molecular Research Center, Ohio, USA) and subsequently reverse transcribed to complementary deoxyribonucleic acid (cDNA) using PrimerScript™ RT Master Mix (Takara, Shiga, Japan) on a SimpliAmp™ Thermal Cycler (Applied Biosystems, Massachusetts, USA) according to the manufacturer’s instructions. Obtained cDNAs were quantified with a reverse transcription quantitative polymerase chain reaction (RT-qPCR) test labeled with SYBR® green (Invitrogen, California, USA) on LightCycler® 96 (Roche, Mannheim, Germany). The gene-specific primers used were as follows: LncRNA LOC100505851 forward, 5'-GCTCTGCAGTGAACACCAAG-3'; LncRNA LOC100505851 reverse, 5'-ACCCCAAATAGCCACCTTGT-3’; β-actin forward, 5'-CATGTACGTTGCTATCCAGGC-3'; β-actin reverse, 5'-CTCCTTAATGTCACGCACGAT-3'; 18s rRNA forward, 5'-GTGTGCCTACCCTACG-3'; and 18s rRNA reverse, 5'-TGACCCGCACTTACTG-3'; U6 forward, 5'-CTCGCTTCGGCAGCACA-3'; U6 reverse, 5'-AACGCTTCACGAATTTGCGT-3'; Gene expression levels were normalized to β-actin/U6 expression using the 2–ΔCt method. Each cDNA sample was triplicated in 96-microwell plates.
Protein-protein interaction (PPI) analysis
The RNA-binding proteins (RBPs) were predicted from starBase (http://starbase.sysu.edu.cn/) and subsequently used to perform PPI network by STRING (https://string-db.org/).
Statistical analyses
The expression of LOC100505851 was calculated as log (lncRNA LOC100505851). Chi-Square test was used to analyze the difference of LOC100505851 expression in cancer and adjacent tissues and the relationship with various clinical pathological characteristics. High LOC100505851 expression was defined as the highest quartile of the expression data set, which was its relative expression ≥0.00186 (pCR analysis set)/0.00205 (correlational analysis set). The associations between all the baseline clinicopathological characteristics [age, tumor (T) stage, nodal (N) stage, estrogen receptor (ER) status, progesterone receptor (PR) status, human epidermal growth factor receptor 2 (HER2) status, Ki-67 index, and molecular subtype based on immunohistochemistry according to St. Gallen Consensus (17) (luminal A-like (ER or PR positive, HER2 negative, Ki-67 <20%) versus luminal B-like (ER or PR positive; HER2 negative, and Ki-67 index ≥20% or HER2 positive and any Ki-67 index) versus HER2 overexpression (HER2 positive, ER and PR negative) versus basal-like (ER and PR negative, HER2 negative)] and lncRNA LOC100505851 expression levels (high versus low) were calculated using the chi-squared test. Gene Expression Profiling Interactive Analysis (GEPIA) online database was used to validate LOC100505851 expression in tumor and normal tissue, as well as its correlation with HER2 expression. pCR was defined as the absence of invasive cancer in the breast and no residual cancer cells in lymph node samples obtained at the time of surgery. Univariate and multivariate logistic regression models were used to evaluate the associations between LOC100505851 expression levels (high versus low) and pCR (adjusted for age, T stage, N stage, ER status, PR status, HER2 status, Ki-67 index).
The receiver operating characteristic (ROC) curves were generated to identify whether LOC100505851 combined with important clinicopathological characteristics: age, T stage, N stage, ER status, PR status, HER2 status, and Ki-67 index. The area under curves (AUC) were compared using the z-test. The estimated median follow-up time was calculated using the reverse Kaplan-Meier method. The Kaplan-Meier plotter (KM plotter) website (http://kmplot.com/analysis/) was used to verify the prognostic value of lncRNA LOC100505851 expression in relapse-free survival (RFS) and OS. Statistical analysis was performed using the statistical software Stata13.0 (Stata Corp LLC, Texas, USA) and the mapping software Graph pad 5.0 (GraphPad Software LLC, California, USA). The results were considered significant when the P value was <0.05.
Results
Baseline characteristics
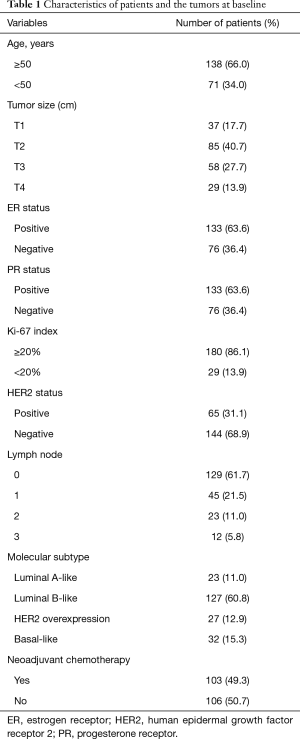
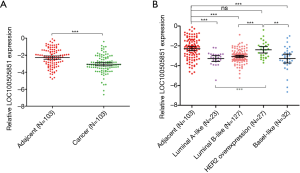
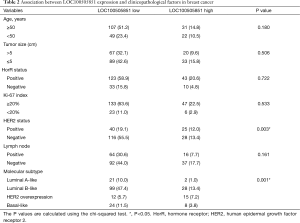
A total of 209 patients were enrolled in the study (Table 1). Of these, 103 received NAT and were included in the pCR analysis. The expression of LOC100505851 in cancer tissues was significantly lower than that in adjacent breast tissue (P<0.001; Figure 1A). The expression of LOC100505851 was significantly lower than that in adjacent breast tissue (Figure 1B) in Luminal A-like (P<0.001), Luminal B like (P<0.001) and basal-like subtypes (P<0.001), but not in HER2 overexpression subtype (P=0.5821). Furthermore, LOC10050585 expression was associated with HER2 expression (P=0.003; Table 2), and it was differentially expressed among the four breast cancer subtypes (luminal A-like, luminal B-like, HER2 overexpression and basal-like) (P<0.001; Figure 1B, Table 2).
Full table
Full table
LncRNA LOC100505851 expression and pCR outcomes
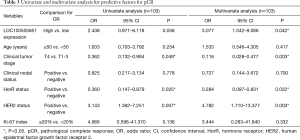
The pCR rate was 30.8% in LOC100505851 low-expression group and 52% in LOC100505851 high-expression group. Clinical tumor stage (OR =0.362, 95% CI: 0.132–0.944, P=0.049), hormone receptor (HorR) status (OR =0.360, 95% CI: 0.147–0.879, P=0.025) and HER2 expression (OR =3.143, 95% CI: 1.362–7.251, P=0.007) were associated with pCR in the univariate logistic analysis (Table 3). In the multivariate analysis, higher LOC100505851 expression was significantly associated with a better pCR rate when adjusted for age, clinical tumor stage, clinical nodal status, HorR status, HER2 status and Ki-67 index (OR =3.077, 95% CI: 1.042–9.086, P=0.042; Table 3). Additionally, clinical tumor stage (OR =0.116, 95% CI: 0.028–0.477, P=0.003), HorR status (OR =0.284, 95% CI: 0.097–0.831, P=0.022) and HER2 expression (OR =4.782, 95% CI: 1.710–13.377, P=0.003) were independently associated with pCR.
Full table
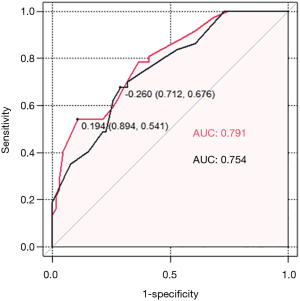
ROC curves were used to evaluate the predictive value of LOC100505851. A higher AUC value was achieved when LOC10050585 was incorporated within the model along with age, clinical T stage, clinical N status, HorR status, HER2 status, and Ki-67 index (AUC 0.791, sensitivity 0.541, specificity 0.894 vs. AUC 0.754, sensitivity 0.676, specificity 0.712; Figure 2).
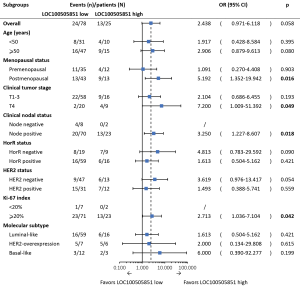
In subgroup analyses, high LOC100505851 expression was associated with a better pCR rate in patients with clinical node-positive disease (OR =3.250, 95% CI: 1.227–8.607, P=0.018), larger tumor size (OR =7.200, 95% CI: 1.009–51.392, P=0.049), higher Ki-67 index (OR =2.713, 95% CI: 1.036–7.104, P=0.042) and postmenopausal status (OR =5.192, 95% CI: 1.352–19.942, P=0.016; Figure 3).
LncRNA LOC100505851 expression and survival
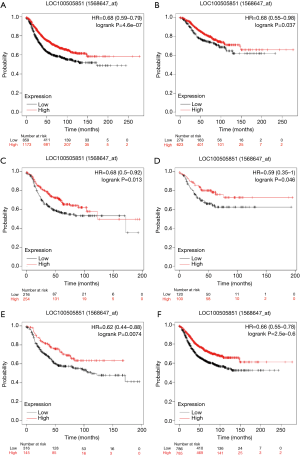
RFS curves were generated with KM plotter. In the overall group, higher LOC100505851 expression was associated with better RFS (HR =0.68, 95% CI: 0.59–0.79, P<0.001; Figure 4A). In the ER positive (HR =0.74, 95% CI: 0.55–0.98, P=0.037; Figure 4B), ER negative (HR =0.68, 95% CI: 0.50–0.92, P=0.013; Figure 4C), triple negative breast cancer (TNBC) (HR =0.59, 95% CI: 0.35–1.00, P=0.046; Figure 4D), HER2 positive (HR =0.62, 95% CI: 0.44–0.88, P=0.0074; Figure 4E) and HER2 negative subgroups (HR =0.66, 95% CI: 0.55–0.78, P<0.001; Figure 4F), higher LOC100505851 expression remained a favorable prognostic marker of RFS. Furthermore, OS curves were generated with KM plotter (Figure S1). In the whole group, higher LOC100505851 expression was associated with better OS (HR =0.60, 95% CI: 0.43–0.84, P=0.0026; Figure S1A). In the ER positive (HR =0.43, 95% CI: 0.22–0.84, P=0.011; Figure S1B), TNBC (HR =0.46, 95% CI: 0.23–0.91, P=0.022; Figure S1D), HER2-positive (HR =0.51, 95% CI: 0.27–0.98, P=0.04; Figure S1E) and HER2 negative subgroups (HR =0.56, 95% CI: 0.39–0.82, P=0.0022; Figure S1F), higher LOC100505851 expression remained a favorable prognostic marker of OS.
Subcellular, PPI network analyses
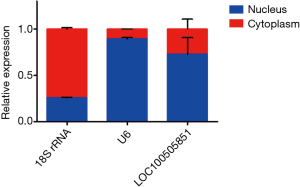
To explore the potential function of LOC100505851, we first determined the subcellular location of LOC100505851. Most of the lncRNA LOC100505851 was located in the nucleus (Figure 5). We further predicted the RBPs and performed subsequent PPI network analyses (Figure 6). We assumed that these RBPs could function by sticking to the scaffold of LOC100505851 and activating multiple signaling pathways.
Discussion
Our study preliminarily explored the expression of lncRNA LOC100505851 in breast cancer and its potential function. For the first time, we observed a positive correlation between LOC100505851 and pCR results and good prognosis in terms of RFS and OS, indicating its clinical value as a novel predictive biomarker for breast cancer patients receiving neoadjuvant chemotherapy and a novel prognostic biomarker for breast cancers.
In our study, we revealed the lower expression of LOC100505851 in tumor tissue than in adjacent tissue. To date, few studies have focused on the role of LOC100505851 in breast cancer. Until recently, Chen and his colleagues searched the Gene Expression Omnibus (GEO) database and retrieved the 177 most differentially expressed genes from the breast cancer expression profile GSE61304. Consistent with our results, lncRNA LOC100505851 was listed among the top 30 lncRNAs as a downregulated gene compared to normal tissue (18). Furthermore, the expression of LOC100505851 in tumor and normal breast tissue in the GEPIA database showed the same pattern as our results (Figure S2). In parallel, the survival analysis with the KM plotter showed that higher LOC100505851 expression was associated with better RFS and OS, which also supported the assumption of LOC100505851 being a tumor suppressor lncRNA. Subcellular localization analysis indicated high expression of LOC100505851 in the nucleus. All of these results suggest a cis role (19) of LOC100505851, as it might recruit transcription factors and affect transcription.
Consistent with the RFS and OS results, LOC100505851 expression was also associated with a greater probability of pCR in the overall patient group, suggesting its potential role as a biomarker in clinical scenarios. More intriguingly, LOC100505851 was associated with HER2 expression (which was validated in the GEPIA database, Figure S3), and furthermore, in both HER2-positive and negative patients, LOC100505851 was associated with a better RFS (Figure 4D) and OS (Figure S1D). Whether this phenomenon was related to drug delivery or clearance in different patients or interactions between the HER2 signaling pathway and LOC100505851 RBPs predicted in Figure 6 awaited further study.
Our work preliminarily revealed the role of LOC100505851 in breast cancer without researching the molecular mechanisms, which was clearly the major limitation of this study. Further study of how LOC100505851 exerts its antitumor function and how it facilitates chemosensitivity is needed.
In conclusion, lncRNA LOC100505851 was significantly expressed at lower levels in breast cancer tissues than in adjacent tissues. Its expression was related to a higher pCR rate, better RFS and better OS, indicating its potential value as a novel positive predictive and prognostic biomarker in breast cancer; however, the mechanism of its function remains to be further explored.
Acknowledgments
Funding: This work was supported by Shanghai Natural Science Foundation (grant numbers 13ZR1452800 and 19ZR1431100), Shanghai Municipal Commission of Health and Family Planning (grant numbers 20144Y0218 and 201640006), Clinical Research Plan of Shanghai Hospital Development Center (grant numbers 16CR3065B, 12016231 and SHDC2020CR3003A) Shanghai ‘Rising Stars of Medical Talent’ Youth Development Program for Outstanding Youth Medical Talents (grant number 2018-16), Shanghai Collaborative Innovation Center for Translational Medicine (grant number TM201908), Multidisciplinary Cross Research Foundation of Shanghai Jiao Tong University (grant numbers YG2017QN49 and ZH2018QNA42), Nurturing Fund of Renji Hospital (grant number PYMDT-002 and PY2018-IIC-01), Science and Technology Commission of Shanghai Municipality (grant number 15JC1402700 and 20DZ2201600), and Shanghai Municipal Key Clinical Specialty.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/gs-21-3
Data Sharing Statement: Available at https://dx.doi.org/10.21037/gs-21-3
Peer Review File: Available at https://dx.doi.org/10.21037/gs-21-3
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/gs-21-3). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study protocols were approved by the ethics committee of Renji Hospital (IRB approval No. 2014-14K and 2017-088). All participants involved in this study provided written informed consents of donating biospecimen for scientific researches. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cardoso F, Spence D, Mertz S, et al. Global analysis of advanced/metastatic breast cancer: Decade report (2005-2015). Breast 2018;39:131-8. [Crossref] [PubMed]
- Cardoso F, Senkus E, Costa A, et al. 4th ESO-ESMO International Consensus Guidelines for Advanced Breast Cancer (ABC 4)†. Ann Oncol 2018;29:1634-57. [Crossref] [PubMed]
- Jones RL, Lakhani SR, Ring AE, et al. Pathological complete response and residual DCIS following neoadjuvant chemotherapy for breast carcinoma. Br J Cancer 2006;94:358-62. [Crossref] [PubMed]
- Cortazar P, Geyer CE Jr. Pathological complete response in neoadjuvant treatment of breast cancer. Ann Surg Oncol 2015;22:1441-6. [Crossref] [PubMed]
- Prowell TM, Pazdur R. Pathological complete response and accelerated drug approval in early breast cancer. N Engl J Med 2012;366:2438-41. [Crossref] [PubMed]
- Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet 2014;384:164-72. [Crossref] [PubMed]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145-66. [Crossref] [PubMed]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47-62. [Crossref] [PubMed]
- Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res 2017;77:3965-81. [Crossref] [PubMed]
- Zhang H, Chen Z, Wang X, et al. Long non-coding RNA: a new player in cancer. J Hematol Oncol 2013;6:37. [Crossref] [PubMed]
- Bolha L, Ravnik-Glavač M, Glavač D. Long Noncoding RNAs as Biomarkers in Cancer. Dis Markers 2017;2017:7243968 [Crossref] [PubMed]
- Strausberg RL, Feingold EA, Grouse LH, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A 2002;99:16899-903. [Crossref] [PubMed]
- Ota T, Suzuki Y, Nishikawa T, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet 2004;36:40-5. [Crossref] [PubMed]
- Jen J, Wang YC. Zinc finger proteins in cancer progression. J Biomed Sci 2016;23:53. [Crossref] [PubMed]
- Zhang H, Sun X, Lu Y, et al. DNA-methylated gene markers for colorectal cancer in TCGA database. Exp Ther Med 2020;19:3042-50. [Crossref] [PubMed]
- Zhou L, Xu S, Yin W, et al. Weekly paclitaxel and cisplatin as neoadjuvant chemotherapy with locally advanced breast cancer: a prospective, single arm, phase II study. Oncotarget 2017;8:79305-14. [Crossref] [PubMed]
- Goldhirsch A, Winer EP, Coates AS, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol 2013;24:2206-23. [Crossref] [PubMed]
- Chen Y, Wu N, Liu L, et al. microRNA-128-3p overexpression inhibits breast cancer stem cell characteristics through suppression of Wnt signalling pathway by down-regulating NEK2. J Cell Mol Med 2020;24:7353-69. [Crossref] [PubMed]
- Chen LL. Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci 2016;41:761-72. [Crossref] [PubMed]



