
KIF23 promotes triple negative breast cancer through activating epithelial-mesenchymal transition
Introduction
Breast cancer is the most common malignant cancer in women all over the world (1), and triple negative breast cancer (TNBC) are highly aggressive with poorer survival rate, in which the expression of estrogen receptor, progesterone receptor and human epidermal growth factor receptor 2 are negative (2). The lack of specific treatment targets makes it difficult to cure TNBC (2), so it is necessary to seek for potential therapeutic targets in TNBC.
KIF23 is a member of kinesin family, recent researches indicate KIF23 plays an important role in the proliferation and migration of malignant cancer cells. KIF23 could promote gastric cancer by stimulating cell proliferation (3) and activate Wnt/β-catenin signaling pathway interacting with Amer1 (4). In lung cancer, KIF23 is overexpressed and identified as an independent prognostic factor (5,6). While knockdown of KIF23 could inhibit the proliferation of pancreatic ductal adenocarcinoma cells (7). In ovarian cancer, KIF23 is identified as a prognostic signature (8). KIF23 is also overexpressed in breast cancer (9), and high expression of KIF23 is significantly correlated with poor overall survival (10). While the function and specific molecule mechanism of KIF23 in TNBC remains unclear.
It is well known that microRNA could combine with target mRNA thus causing mRNA degradation, recent researches reveal that miR-424-5p and miR-503-5p could directly targeted to 3'UTR of KIF23 in ovarian cancer (11) and human glioma (12). Similarly, in prostate cancer, KIF23 is targeted by miR-15a-5p (13). While, miRNAs that target KIF23 mRNA in TNBC is still unknown.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/gs-21-19).
Methods
Tissue samples
30 cases of TNBC tissues and paired paracancer tissues were collected from the Department of Breast and Thyroid Surgery of Shanghai Tenth People’s Hospital The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of Shanghai Tenth People’s Hospital (NO. 2020-KN174-01). All patients were contacted by telephone to obtain verbal informed consent. Tissues applied to qPCR were cryopreserved in liquid nitrogen following tissue samples separated from breast cancer tissues without influencing pathologic diagnosis. On condition that cancer tissues were diagnosed as TNBC, tissues were included in this research. Specimens used for immunohistochemistry were gathered from the Department of Pathology of Shanghai Tenth People’s Hospital.
Immunohistochemistry
Tissues applied to immunohistochemistry were stored in formalin then embedded with paraffin. Firstly, deparaffinizing and rehydrating the paraffin section, then retrieving tissue antigen with citric acid (pH 6.0). Secondly, blocking endogenous peroxidase activity with 3% hydrogen peroxide and sealing with 3% BSA. Thirdly, primary antibody and secondary antibody were applied in turn for incubation. After diaminobenzidine (DAB) chromogenic reaction, nucleus counterstaining, dehydration and mounting, illustrative images were gathered under a microscope.
Cell culture
Human normal breast epithelial cell line MCF10A, TNBC cell lines MDA-MB-231 and BT549, and human embryonic kidney cell line 293T were purchased from the Chinese Academy of Science, Shanghai. DMEM medium with high Glucose (Gibco, 11965092, USA) was used for cell culture of MDA-MB-231 and 293T. RPMI-1640 medium (Gibco, 21870076, USA) was used for cell culture of BT549. MCF10A cells were cultured in Mammary Epithelial Cell Medium (Procell, CM-0525, China). All mediums were added with 10% Fetal Bovine Serum (FBS; Gibco, 10099141C, USA), along with penicillin (100 U/mL) and streptomycin (100 µg/mL) (PS; Servicebio, G4003, China). 0.25% Trypsin-EDTA Solution (Servicebio, G4004, China) was applied for cell passage cultivation. 37 °C with 5% CO2 cell incubator was applied for cell incubation.
Cell transfection
In order to change the expression of KIF23 in TNBC cells, siRNA targeting KIF23 was applied. TNBC cell lines MDA-MB-231 and BT549 were transfected with Lipofectamine®3000 (Invitrogen, L3000-015, USA). KIF23-siRNA (KIF23-si1 # Sense: 5'-GACUAUAUCUAGAUCAUGUCU-3', anti-sense: 5'-ACAUGAUCUAGAUAUAGUCUU-3'; KIF23-si2 #sense: 5'-GAAGUGAUCAAUAAUACAACU-3', anti-sense: 5'-UUGUAUUAUUGAUCACUUCUA-3'; KIF23-si3 #sense: 5'-GAGACUCAGUAUUCAUUUAAA-3', anti-sense: 5'-UAAAUGAAUACUGAGUCUCCU-3') or negative control (NC #sense: 5'-UUCUCCGAACGUGUCACGUTT-3', antisense: 5'-ACGUGACACGUUCGGAGAATT-3') sequence were synthesized by GENEray (Shanghai, China). Three time points (24 h, 48 h, 72 h) were set to compare the knockdown efficiency.
Quantitative real-time PCR (qPCR)
Total RNA was extracted from MCF10A, MDA-MB-231, BT549 cells, TNBC tissues and paired paracancer tissues with TRIzol Reagent (Invitrogen, 15596026, USA), 1000ng total RNA was used for first strand cDNA synthesis according to protocol of Hifair® III 1st Strand cDNA Synthesis SuperMix for qPCR (YEASEN, 11141ES10, China), qPCR was performed using Hieff UNICON® qPCR SYBR Green Master Mix (YEASEN, 11200ES03, China). GAPDH (forward primer: 5'-GGAGCGAGATCCCTCCAAAAT-3' and reverse primer: 5'-GGCTGTTGTCATACTTCTCATGG-3') was used for normalized expression of KIF23 (forward primer: 5'-AGTCAGCGAGAGCTAAGACAC-3' and reverse primer: 5'-GGTTGAGTCTGTAGCCCTCAG-3'), primers applied for qPCR were synthesized by GENEray (Shanghai, China). 2−ΔΔCt method was applied to analyze expression of the indicated mRNA.
CCK-8 assay
For CCK-8 assay, transfected MDA-MB-231 and BT549 cells were plated in 96-well plates at a density of 1,000 cells per well, cell viability was assessed at 0 h, 24 h, 48 h, 72 h and 96 h. According to the manufacturer’s instructions of CCK-8 (YEASEN, 40203ES60, China), 100 µL complete medium was used for cell incubation, 10 µL CCK-8 per well was added at set time point, after 3 hours incubation in 37 °C with 5% CO2, the absorbance value was quantified at a wavelength of 450 nm.
Colony formation assay
For colony formation assay, transfected MDA-MB-231 and BT549 cells were plated in 6-well plates at a density of 500 cells per well, and cells were incubated for 7–14 days at 37 °C with 5% CO2, medium was changed every 3 days. Following observation of clones with naked eyes, medium were removed and cells were washed with phosphate buffer saline (PBS, Servicebio, G4202-500ML, China) for three times and fixed with 95% ethanol for 10 minutes and removed, finally clones were stained with 0.1% crystal violet (YEASEN, 60506ES60, China).
Wound healing assay
MDA-MB-231 and BT549 cells were seeded in 6-well plates at a density of 50–60% for the first day, 20 hours later, transfected cells with KIF23-siRNA or negative control, incubated cells with complete medium at 37 °C with 5% CO2, scratch was made using pipette tips when cell density reached 90–95%, washed cells with PBS and incubated cells with DMEM/ RPMI-1640 medium supplemented with 2% FBS. Recorded the wound healing with light microscope at 0 h, 24 h and 48 h after scratching. Wound Closure % = [(A0 − At)/A0] ×100% (A0 is the area of the wound measured immediately after scratching, At is the area of the wound measured t hours after the scratch is made).
Transwell assay
To explore the change of migration ability after KIF23 knockdown in MDA-MB-231 and BT549, 5×104 transfected cells were plated in upper chamber of transwell (Corning, 3422, USA) incubated with serum free medium, and medium supplemented with 10% FBS was added to the lower chamber, 18 hours later, cells moved to lower chamber of transwell were fixed by 4% paraformaldehyde and stained with 0.05% crystal violet. Representative images were collected under light microscope.
Western blot analysis
Protein was extracted with RIPA lysis buffer (YEASEN, 20101ES60, China) and concentration was measured by BCA Protein Quantification Kit (YEASEN, 20201ES76, China). 30 µg total protein was applied for polyacrylamide gel electrophoresis (PAGE), and proteins separated in gel were transferred to nitrocellulose membrane (Pall, 40038684, USA). Following transfer, the membrane was blocked with 5% non-fat milk for 1 hour and, incubated with primary antibodies of KIF23 (absin, abs132952, China), E-Cadherin (ABclonal, A3044, China), N-Cadherin (ABclonal, A19083, China), Vimentin (ABclonal, A19607, China), and GAPDH (Servicebio, GB11002, China) for 12–18 hours in 4 °C. The membrane was then washed with Tris Buffered saline Tween for three times following which, membranes were incubated with the secondary antibody IRDye® 800CW Goat anti-Rabbit IgG (LI-COR, USA) in room temperature for 1 hour. The immunoreactive protein bands on the membrane were scanned with Odyssey scanning system (Li-COR, USA).
Dual luciferase reporter assay
293T cells were divided into 4 groups, group 1: transfected 293T cells with miRNA mimics and wildtype KIF23-3’UTR plasmid; group 2: transfected with negative control mimics and mutated KIF23-3’UTR plasmid; group3: transfected with miRNA mimics and mutated KIF23-3’UTR plasmid; group 4: transfected with negative control mimics and mutated KIF23-3’UTR plasmid. 48 hours later, the activity of firefly luciferase and renilla luciferase were detected according to the protocol for Dual Luciferase Reporter Gene Assay Kit (Promega, E1910, USA).
Statistical analysis
Data was presented as mean ± standard deviation (SD) and analyzed by SPSS 22.0 or GraphPad Prism 9.0. One-way ANOVAs, Student’s t-test or χ2 test were applied to compare difference between groups. P values <0.05 was considered as significantly different.
Results
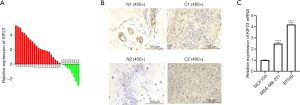
KIF23 is overexpressed in triple negative breast cancer tissues compared to paired paracancer tissues
According to the UALCAN database (http://ualcan.path.uab.edu/) (14) data, KIF23 expression was higher in breast invasive ductal carcinoma than that of normal breast tissue (Figure S1A). Nextly, the expression of KIF23 mRNA was detected by qPCR in 30 paired TNBC tissues, and the results revealed that KIF23 was significantly increased in TNBC tissues compared to paired paracancer tissues (Figure 1A). To verify the specificity of KIF23 primer, the dissociation curve was analyzed in qPCR. As our result showed (Figure S1B), the single peak curve indicated primer of KIF23 in our research was specific. We also confirmed the expression of KIF23 protein in TNBC tissues through immunohistochemistry, in which expression of KIF23 protein was also increased in most TNBC tissues (Figure 1B). Meanwhile, we analyzed expression of KIF23 mRNA in MCF10A, MDA-MB-231 and BT549, the results showed expression of KIF23 was distinctly increased in TNBC cell lines (Figure 1C).
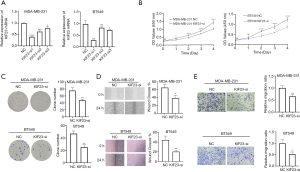
SiRNA-induced knockdown of KIF23 inhibits proliferation, colony formation and migration of triple negative breast cancer cells
In order to explore function of KIF23 in triple negative breast cancer, we decreased expression of KIF23 via siRNA targeting KIF23 in MDA-MB-231 and BT549, qRT-PCR and western blot analysis were carried out to confirm knockdown efficiency of KIF23. KIF23-si1 (sense: 5’-GACUAUAUCUAGAUCAUGUCU-3’, anti-sense: 5’-ACAUGAUCUAGAUAUAGUCUU-3’) shows the highest knockdown efficiency (Figure 2A), thus we transfected MDA-MB-231 and BT549 with KIF23-si1 to observe proliferation, colony formation and migration of triple negative breast cancer cells. CCK-8 assay showed knockdown of KIF23 significantly suppressed proliferation of TNBC cells (Figure 2B), similarly, colony formation assay revealed downregulation of KIF23 decreased colony formation of MDA-MB-231 and BT549 (Figure 2C). To predict migration of TNBC cells, we carried out wound healing experiments and transwell assays, inhibition of KIF23 restrained migration of MDA-MB-231 and BT549 (Figure 2D,E). These results indicated that knockdown of KIF23 leads to decrease in cell proliferation, colony formation as well as cell migration of TNBC
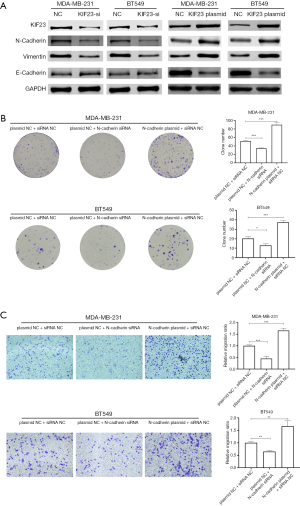
Suppression of KIF23 inhibits Epithelial-mesenchymal transition (EMT)
Since KIF23 showed significant influence on proliferation and migration of TNBC cells, we were eager to explore the specific molecular mechanisms of KIF23 in TNBC cells. EMT plays important role in proliferation and migration of breast cancer cells. Hence, we inhibited KIF23 by siRNA and detected expression change of KIF23, E-Cadherin, N-Cadherin, and Vimentin. As our results showed, after knockdown of KIF23, expression of KIF23, N-Cadherin and Vimentin decreased, and expression of E-cadherin increased, while overexpression of KIF23 in TNBC cells increased expression of N-cadherin and Vimentin, and expression of E-cadherin decreased (Figure 3A), which indicated that suppression of KIF23 inhibited EMT and activation of KIF23 promotes EMT. Meanwhile, knockdown of N-cadherin suppressed proliferation and migration of TNBC cells, and overexpression of N-cadherin promotes proliferation and migration of TNBC cells (Figure 3B,C).
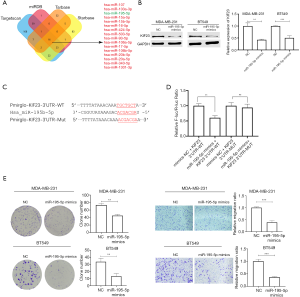
Hsa-miR-195-5p combine with 3’UTR of KIF23 mRNA thus promoting degradation of KIF23
MicroRNAs are kind of non-coding RNAs which mainly combine with 3’UTR of target mRNA, hence promoting degradation of target mRNA. Firstly, we collected potential upstream microRNA targeting KIF23 mRNA from starbase database (http://starbase.sysu.edu.cn/) (15), targetscan database (http://www.targetscan.org/) (16), miRDB database (http://mirdb.org/) (17) and Tarbase database (http://carolina.imis.athena-innovation.gr/) (18). Resultsfrom the sources presented 16 microRNAs were potential to regulate KIF23 in breast cancer (Figure 4A). Hsa_miR-195-5pwas a potential tumor suppressor in breast cancer in our previous study (19), expression of miR-195-5p was lower in breast cancer tissues and negative correlated with expression of KIF23 mRNA according to the starbase database (Figure S1C,D). Next, we transfected MDA-MB-231 and BT549 with hsa-miR-195-5p mimics to detect expression change of KIF23, interestingly, similar to siRNA-induced downregulation of KIF23, after TNBC cells transfected with miR-195-5p mimics, expression of KIF23 decreased (Figure 4B). To further explore the relationship between hsa-miR-195-5p and KIF23, we carried out dual luciferase reporter assay. 293T cells were transfected with miR-195-5p mimics and wildtype KIF23-3’UTR or mutated KIF23-3’UTR. Co-transfected 293T cells with hsa-miR-195-5p mimics and wildtype KIF23-3’UTR group have the lowest luciferase activity (Figure 4C,D), which indicated that miR-195-5p directly combines with 3’UTR of KIF23 mRNA. To explore biological function of miR-195-5p in TNBC cells, we transfected miR-195-5p mimics into MDA-MB-231 and BT549 cells, colony formation assays and transwell assays revealed miR-195-5p could inhibit proliferation and migration of TNBC cells (Figure 4E).
Discussion
Triple negative breast cancer is the most challenging breast cancer subtype with higher rates of relapse and metastatic potential compared to other breast cancer subtypes. In early TNBC, patients could benefit from pembrolizumab (PD-L1 antagonistic monoclonal antibody) plus neoadjuvant chemotherapy (20). BRCA1/2 are well-studied gene in TNBC, and it is encouraging that the adenosine diphosphate-ribose polymerase inhibitor Olaparib could be effective in TNBC patients with BRCA mutation (21). However, because of the intrinsic complexity of TNBC, it is necessary to find out novel therapeutic targets. Kinesin superfamily proteins are involved in cells mitosis, meiosis, and the transport of macromolecules, aberrant expression of Kinesins could result in cell death, chromosome duplication tumorigenesis and tumor progression (22).
In our study, KIF23 was overexpressed in TNBC tissues compared to paired paracancer tissues, which was consistent with previous study (9). After transfection with KIF23-siRNA or negative control in MDA-MB-231 and BT549, CCK-8 assay, colony formation, wound healing assay and transwell assay were carried out to explore phenotypic change. As our results showed, knockdown of KIF23 by siRNA in TNBC cell lines MDA-MB-231 and BT-549 results in inhibition of proliferation and migration. Our data preliminarily showed that KIF23 plays an oncogene role in TNBC.
Epithelial-mesenchymal transition (EMT) is a biological process by which epithelial cells loss expression of E-cadherin and acquire the ability to migrate (23). Knockdown of KIF23 suppresses the expression of N-Cadherin and Vimentin, while expression of E- Cadherin is increased. While the exact mechanism how KIF23 influence EMT is still unclear, further studies should be conducted.
MicroRNAs are cluster of small non-coding RNAs mainly combine with 5’-UTR or 3’UTR of target mRNA thus promoting degradation of mRNA. We analysis 4 databases and find miR-195-5p as a potential upstream miRNA targeting KIF23 mRNA, qRT-PCR results revealed that miR-195-5p is downregulated in TNBC tissues. Hence, we carried out dual luciferase reporter assay and confirmed miR-195-5p could combine with 3’UTR of KIF23 mRNA.
In conclusion, KIF23 is overexpressed in TNBC tissues, siRNA induced knockdown of KIF23 inhibits proliferation and migration of MDA-MB-231 and BT-549 cells. MiR-195-5P is downregulated in TNBC tissues and suppress expression of KIF23 by directly combining with 3’UTR of KIF23 mRNA. Our results provide a potential novel target to TNBC.
Acknowledgments
We give special thanks to all other members in our research group for their valuable suggestion.
Funding: This work was supported by the National Natural Science Foundation of China (No. 82073204) and Shanghai Municipal Health Commission (No. 202040157).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/gs-21-19
Data Sharing Statement: Available at https://dx.doi.org/10.21037/gs-21-19
Peer Review File: Available at https://dx.doi.org/10.21037/gs-21-19
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/gs-21-19). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of Shanghai Tenth People’s Hospital (NO. 2020-KN174-01). All patients were contacted by telephone to obtain verbal informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Harbeck N, Penault-Llorca F, Cortes J, et al. Breast cancer. Nat Rev Dis Primers 2019;5:66. [Crossref] [PubMed]
- Garrido-Castro A C, Lin N U, Polyak K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov 2019;9:176-198. [Crossref] [PubMed]
- Li X L, Ji Y M, Song R, et al. KIF23 Promotes Gastric Cancer by Stimulating Cell Proliferation. Dis Markers 2019;2019:9751923 [Crossref] [PubMed]
- Liu Y, Chen H, Dong P, et al. KIF23 activated Wnt/β-catenin signaling pathway through direct interaction with Amer1 in gastric cancer. Aging (Albany NY) 2020;12:8372-96. [Crossref] [PubMed]
- Kato T, Wada H, Patel P, et al. Overexpression of KIF23 predicts clinical outcome in primary lung cancer patients. Lung Cancer 2016;92:53-61. [Crossref] [PubMed]
- Vikberg A L, Vooder T, Lokk K, et al. Mutation analysis and copy number alterations of KIF23 in non-small-cell lung cancer exhibiting KIF23 over-expression. Onco Targets Ther 2017;10:4969-79. [Crossref] [PubMed]
- Gao C T, Ren J, Yu J, et al. KIF23 enhances cell proliferation in pancreatic ductal adenocarcinoma and is a potent therapeutic target. Ann Transl Med 2020;8:1394. [Crossref] [PubMed]
- Hu Y, Zheng M, Wang C, et al. Identification of KIF23 as a prognostic signature for ovarian cancer based on large-scale sampling and clinical validation. Am J Transl Res 2020;12:4955-76. [PubMed]
- Li T F, Zeng H J, Shan Z, et al. Overexpression of kinesin superfamily members as prognostic biomarkers of breast cancer. Cancer Cell Int 2020;20:123. [Crossref] [PubMed]
- Song X, Zhang T, Wang X, et al. Distinct Diagnostic and Prognostic Values of Kinesin Family Member Genes Expression in Patients with Breast Cancer. Med Sci Monit 2018;24:9442-64. [Crossref] [PubMed]
- Li T, Li Y, Gan Y, et al. Methylation-mediated repression of MiR-424/503 cluster promotes proliferation and migration of ovarian cancer cells through targeting the hub gene KIF23. Cell Cycle 2019;18:1601-18. [Crossref] [PubMed]
- Zhao C, Wang XB, Zhang YH, et al. MicroRNA-424 inhibits cell migration, invasion and epithelial-mesenchymal transition in human glioma by targeting KIF23 and functions as a novel prognostic predictor. Eur Rev Med Pharmacol Sci 2018;22:6369-78. [PubMed]
- Wu H, Tian X, Zhu C. Knockdown of lncRNA PVT1 inhibits prostate cancer progression in vitro and in vivo by the suppression of KIF23 through stimulating miR-15a-5p. Cancer Cell Int 2020;20:283. [Crossref] [PubMed]
- Chandrashekar D S, Bashel B, Balasubramanya S A H, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Li J H, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42:D92-7. [Crossref] [PubMed]
- Agarwal V, Bell G W, Nam J W, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4:e05005 [Crossref] [PubMed]
- Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res 2020;48:D127-D131. [Crossref] [PubMed]
- Karagkouni D, Paraskevopoulou M D, Chatzopoulos S, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res 2018;46:D239-D245. [Crossref] [PubMed]
- Luo Q, Wei C, Li X, et al. MicroRNA-195-5p is a potential diagnostic and therapeutic target for breast cancer. Oncol Rep 2014;31:1096-102. [Crossref] [PubMed]
- Schmid P, Cortes J, Pusztai L, et al. Pembrolizumab for Early Triple-Negative Breast Cancer. N Engl J Med 2020;382:810-21. [Crossref] [PubMed]
- Robson M, Im S A, Senkus E, et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N Engl J Med 2017;377:523-33. [Crossref] [PubMed]
- Lucanus A J, Yip G W. Kinesin superfamily: roles in breast cancer, patient prognosis and therapeutics. Oncogene 2018;37:833-8. [Crossref] [PubMed]
- Dongre A, Rashidian M, Eaton E N, et al. Direct and Indirect Regulators of Epithelial-Mesenchymal Transition (EMT)-mediated Immunosuppression in Breast Carcinomas. Cancer Discov 2021;11:1286-305. [Crossref] [PubMed]


