
An immunohistochemical panel of three small ubiquitin-like modifier genes predicts outcomes of patients with triple-negative breast cancer
Introduction
Triple-negative breast cancer (TNBC) is a specific type of breast cancer that is characterized by the absence of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor-2 (HER2) expression. Although TNBC only constitutes approximately 10–15% of all breast cancers (1,2), it has a highly aggressive clinicopathological signature and unfavorable outcomes (3). Patients with TNBC generally develop distant metastasis within the first three years after initial treatment, with the mortality rate reaching about 40% in the first five years (4). The lack of ER/PR and HER2 expression renders TNBC inaccessible to endocrine or anti-HER2 target therapies. Therefore, the most common treatment strategy for TNBC is a combination of surgery, chemotherapy, and radiotherapy. At present, anthracycline- and taxane-based adjuvant chemotherapy is the standard regimen for TNBC patients after resection (5,6). TNBC has a high sensitivity to chemotherapy, for patients with TNBC have an enhanced neoadjuvant response rate compared with other subtypes of breast cancer (7,8). However, some patients still develop a rapid onset of recurrence and poor prognosis, which is commonly referred to as the “triple-negative paradox” (9). Thus, identification of new predictive biomarkers for chemotherapy response and promising therapeutic targets might be beneficial in the treatment of TNBC.
As an important post-translational protein modification, small ubiquitin-like modifiers (SUMOs) mediate post-translational modifications (SUMOylation) has attracted increasing attention. Four subtypes of SUMO have been identified, including SUMO1, SUMO2, SUMO3, and SUMO4 (10). SUMO2 and SUMO3 are 95% identical to each other and only 50% identical to SUMO1 (11). SUMO1/2/3 are ubiquitously expressed in human tissues, however SUMO4 is only expressed in spleen lymph nodes and the kidney (12). SUMOylation is catalyzed by a three-step enzymatic reaction, including activation, coupling, and ligation (13). SUMO E1-activating enzyme is a protein that contains two subunits, namely, SUMO-activating enzyme E1 (SAE1) and SUMO-activating enzyme E2 (SAE2). Ubiquitin-Conjugating Enzyme 9 (UBC9) is the only known SUMO-conjugating E2 enzyme required for SUMOylation, and its deletion abolishes SUMO conjugation (14). SUMO E3 ligases are roughly divided into three categories including the protein inhibitor of activated signal transducer and activator of transcription 1 (STAT 1) protein family, the nucleoporin Ran binding protein 2, and the human polycomb protein Pc2. Although SUMO is similar to ubiquitin, SUMOylation does not directly lead to protein degradation, but leads to the regulation of cell functions, such as protein-protein interactions, maintenance of genome integrity, subcellular localization, transcription regulation, deoxyribonucleic acid (DNA) repair, and cell cycle (11,15). The dysregulation of SUMOylation could result in tumor progression, and is considered as a novel biomarker and possible therapeutic target for cancers (16). For instance, one previous study reported that the expression of SUMO E3 ligase PIAS1 could serve as a useful prognostic biomarker in patients with breast cancer (17). However, no studies to date have focused on the expression and prognostic value of SUMO1/2/3.
In this study, we sought to identify the expression and prognostic utility of SUMOs and aimed to build a prognosis prediction model based on SUMO1/2/3 protein expression. Potential mechanisms that regulate the SUMOylation pathway in TNBC were also explored. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/gs-21-37).
Methods
Extraction of gene expression data from TNBC patient datasets
The microarray datasets of TNBC patients were extracted from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) and the Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). Four microarray gene expression datasets of TNBC patients (GSE31448, GSE45827, GSE53752, and GSE65216) were obtained from the GEO database. The method for extracting microarray gene expression values was based on our previous research (18-20).
Patients and specimens
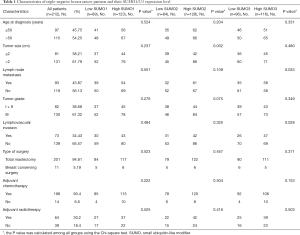
A total of 212 TNBC patients from Fujian Medical University Union Hospital between June 2013 and August 2017 were retrospectively reviewed. The included patients had a median age of 51 years (range, 27–77 years), histologically confirmed TNBC, as well as 4–77 months of follow-up data. Clinicopathological information, including age, tumor size, nodal status, tumor grade, lymphovascular invasion (LVI), type of surgery, chemotherapy and radiotherapy status, were obtained from medical records. Disease-free survival (DFS) was defined as the time from the date of diagnosis to the date of clinical relapse (with histopathology confirmation or radiological evidence of tumor recurrence). Overall survival (OS) was defined as the time from the date of diagnosis until death from any cause. The follow-up deadline was August 30, 2020.
The inclusion criteria were as follows: (I) patients with no history of other malignant tumors, bilateral breast cancer, or de novo IV stage; (II) patients who received total mastectomy or breast conserving surgery without neoadjuvant chemotherapy or radiotherapy; (III) primary tumor size was pT1c-pT2 (1 cm < T ≤5 cm); and (IV) demographic, clinicopathological, and follow-up information were complete. Patients who had received at least three cycles of anthracycline-based and three cycles of taxane-based regimens were considered as having chemotherapy, while those with insufficient chemotherapy cycles were excluded from the study.
This study was approved by the Research Ethics Committee of Fujian Medical University Union Hospital (2019KJCX011). Informed consent was obtained from each participant. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Immunohistochemistry (IHC) staining analysis
IHC staining analysis was performed to measure the protein expression of SUMO1/2/3 in all TNBC tissues and adjacent normal breast tissues according to the standard immunoperoxidase staining procedure. Slides were incubated with anti-SUMO1 (ab32058, Abcam, Cambridge, UK, diluted 1:150), anti-SUMO2 (ab233222, Abcam, Cambridge, UK, diluted 1:300) and anti-SUMO3 (ab203570, Abcam, Cambridge, UK, diluted 1:300) according to the manufacturer’s instructions. To ensure quality, a negative control was prepared via substitution of a primary antibody with phosphate-buffered saline (PBS).
The IHC staining scores of SUMO1/2/3 were assessed by two independent pathologists. The percentage of stained positive cells was scored from 1 to 4: 1, 0–25%; 2, 26–50%; 3, 51–75%; and 4, 75–100%. The staining intensity score was calculated from 0 to 3: 0, no staining; 1, weak staining; 2, moderate staining; and 3, strong staining. The final scores were based on the sum of these two scores. A score >5 was defined as high expression level and a score ≤5 was defined as low expression.
Gene set variation analysis (GSVA) and LASSO analysis
GSVA provides increased power to detect subtle pathway activity changes in a sample population compared to corresponding methods. In this study, the pathway activity of protein SUMOylation and 50 oncogene pathways in TNBC were analyzed. The GSVA analysis was performed using R package ‘GSVA’. We used the Least absolute shrinkage and selection operator (LASSO) Cox regression model to construct a three SUMOs-based classifier (SB classifier) for predicting the DFS of TNBC patients. The LASSO analysis was performed using R package ‘glmnet’.
Statistical analysis
In this study, the t-test was used to compare continuous variables in two groups. Correlations between SUMO1/2/3 expression and clinicopathological characteristics were identified by the chi-squared test. DFS and OS were calculated by the Kaplan-Meier method and differences between groups were examined with the log-rank test. We performed Cox regression analysis to undertake univariate and multivariate survival analyses. All P values <0.05 were considered statistically significant.
Results
Protein SUMOlyation pathway was activated in TNBC
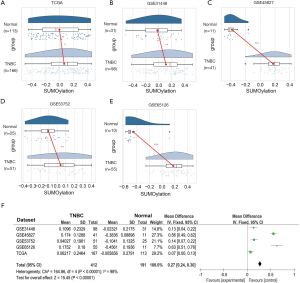
In order to explore the pathway activity of protein SUMOlyation in TNBC, TCGA database and four related GEO databases (GSE31448, GSE45827, GSE53752, and GSE65216) were employed. TCGA database contained 166 cases of TNBC tissues and 113 adjacent normal breast tissues. As for the four GEO databases, 98 cases of TNBC tissues and 31 adjacent normal breast tissues were from the GSE31448 database, 41 cases of TNBC tissues and 11 adjacent normal breast tissues were from the GSE45827 database, the GSE53752 database consisted of 51 cases of TNBC tissues and 25 adjacent normal breast tissues, while 55 cases of TNBC tissues and 10 adjacent normal breast tissues were retrieved from the GSE65216 database. GSVA was performed to conduct Gene Ontology (GO) analysis and assign protein SUMOlyation pathway activity estimates to individual samples from TCGA and GEO databases.
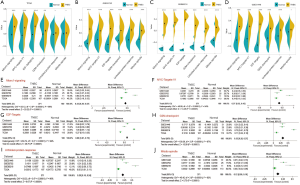
It was found that the protein SUMOlyation pathway exhibited a higher enrichment score in the TNBC tissues compared with adjacent normal breast tissues (GSE45827, GSE65216, P<0.001; GSE31448, GSE53752, P<0.01; TCGA, P<0.05) (Figure 1A,B,C,D,E). Moreover, meta-analysis containing 603 tissues from five TNBC databases mentioned above further demonstrated that protein SUMOlyation pathway was activated in TNBC (P<0.001; Figure 1F).
High SUMO1/2/3 protein expression were unfavorable prognostic factors for TNBC patients
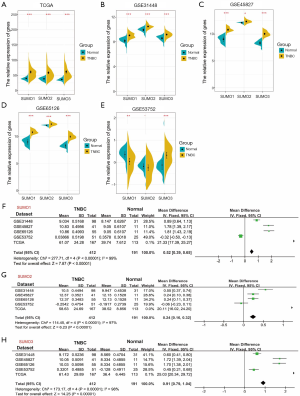
To investigate whether SUMO1, SUMO2, and SUMO3 were involved in TNBC progression, we analyzed their expression in the TCGA and four GEO databases. SUMO1, SUMO2, and SUMO3 were up-regulated in TNBC tissues compared with adjacent normal breast tissues in TCGA, GSE31448, GSE45827, and GSE65216 databases (Figure 2A,B,C,D). As for the GSE53752 database, up-regulation of SUMO3 expression and down-regulation of SUMO1 and SUMO2 in TNBC tissues compared with adjacent normal breast tissues were observed (SUMO1, P<0.01; SUMO2, P>0.05; SUMO3, P<0.001) (Figure 2E). Remarkably, meta-analysis revealed that the mRNA expression of SUMO1, SUMO2, and SUMO3 were increased in TNBC (Figure 2F,G,H).
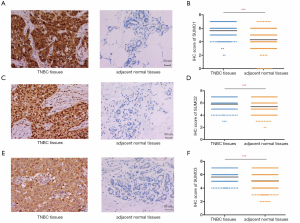
In order to further validate the data from TCGA and GEO databases, we performed an IHC study with patient samples to determine the protein expression of SUMO1, SUMO2, and SUMO3 in TNBC. Immunohistochemistry analysis revealed that SUMO1, SUMO2, and SUMO3 were significantly up-regulated in 212 TNBC tissues compared to the paired adjacent normal breast tissues (Figure 3). The clinicopathological characteristics of patients in the study cohort are summarized in Table 1. Most of the patients (93.4%) were treated with adjuvant chemotherapy. We estimated the correlations of SUMO1/2/3 expression with relevant clinicopathological factors. No associations between SUMO1 expression and clinicopathological features were observed. SUMO2 expression was indicated to be significantly associated with tumor size (P=0.032), while SUMO3 expression was significantly correlated with lymph node metastasis (P=0.033) and lymphovascular invasion (P=0.028).
Full table
Survival analysis was conducted to explore the relationship between SUMO1/2/3 protein expression, clinicopathological factors, and survival of these 212 TNBC patients. Kaplan-Meier analysis for OS and DFS of TNBC patients was performed according to SUMO1/2/3 protein expression (Figure 4A,B,C,D,E,F), which implied that TNBC patients with higher expression of SUMO1/2/3 suffered a lower OS (Figure 4A,B,C) and DFS (Figure 4D,E,F). Univariate and multivariate Cox regression analyses were conducted to clarify the independent factors affecting OS and DFS of TNBC patients.
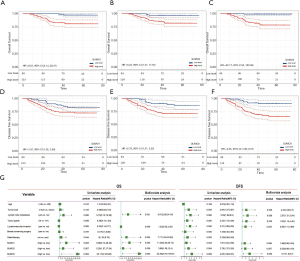
In order to identify the independent factors impacting patient outcome, univariate and multivariate Cox analyses were performed. Lymph node metastasis, radiotherapy, as well as SUMO1, SUMO2, and SUMO3 protein expression were finally determined to be independent prognostic factors for OS of TNBC patients by multivariate Cox analyses (Figure 4G). As for DFS, tumor size, lymph node metastasis, tumor grade, lymphovascular invasion, and SUMO3 protein expression were determined to be independent prognostic factors in TNBC patients (Figure 4G).
Construction of a prognostic scoring model based on SUMO1/2/3 proteins
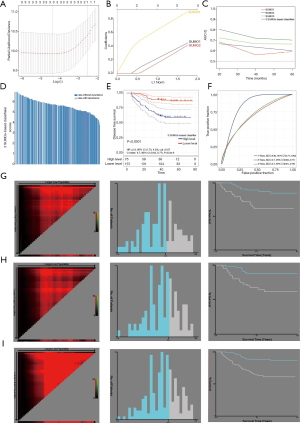
In order to construct a risk score model for predicting the DFS of TNBC, we constructed a LASSO Cox regression model to build a SUMO proteins-based prognostic classifier, which included SUMO1, SUMO2, and SUMO3, and called it the ‘SB classifier’ (Figure 5A,B). Using LASSO Cox regression models, we calculated a risk score for each patient based on individualized values of IHC scores for the three proteins: Risk score = (SUMO1 × 0.3746) + (SUMO2 × 0.3290) + (SUMO3 × 0.8217). The SB classifier possessed significantly higher prognostic accuracy than a single SUMO alone (Figure 5C). When we assessed the distribution of risk scores and recurrence status, TNBC patients with higher risk scores generally had a higher recurrence rate than those with lower risk scores (Figure 5D). TNBC patients were then assigned into a SB classifier high-level group (75 patients) and low-level group (137 patients) by the cut-off value (5.87). The Kaplan-Meier curve showed that patients in the SB classifier high-level group presented a significantly worse DFS [hazard ratio (HR) 2.8, 95% confidence interval (CI): 1.73–4.53, P<0.01] (Figure 5E). By predicting the DFS of TNBC patients at 1, 3, and 5 years, the areas under the receiver operating characteristic (ROC) curves (AUC) obtained from the risk-based prediction model were 0.84, 0.7, and 0.7, respectively (Figure 5F). The total cohort was randomly divided into two equal training and validation sets using X-tile plots. Based on cut-points of the risk score, TNBC patients were divided into SB classifier low-level and SB classifier high-level in the training cohort. Patients with poor DFS exhibited a higher risk score than those with good prognosis (Figure 5G). Similar outcomes were observed in the validation and total cohorts (Figure 5H,I).
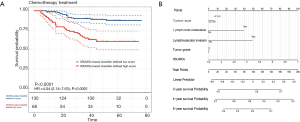
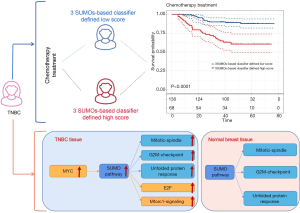
Survival analysis based on our SB classifier showed that patients in the classifier-defined low score group had a favourable response to chemotherapy (HR 4.04, 2.14–7.63; P<0.0001) (Figure 6A), which indicated that our SB classifier could successfully identify patients with TNBC who might benefit from chemotherapy. To provide clinicians with a quantitative method to predict the probability of disease recurrence in TNBC patients undergoing chemotherapy, we constructed a nomogram that integrated both the SB classifier and clinicopathological factors (Figure 6B).
Oncogenic pathways that positively correlate to protein SUMOylation were activated in the tumors of TNBC patients
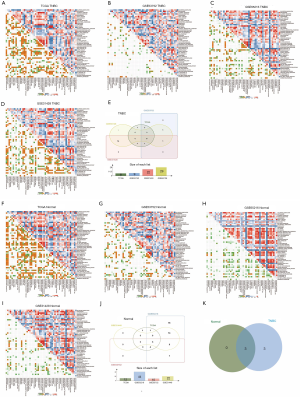
Using GSVA method and the Molecular Signatures Database hallmark gene set collection, we analyzed the mRNA expression data of TNBC in the TCGA, GSE53752, GSE65216, and GSE31448 databases. The correlation between protein SUMOylation and 50 hallmark gene set in TNBC was analyzed by Pearson correlation analysis. In the tumor samples of the TNBC cohort, the intersection of TCGA, GSE53752, GSE65216, and GSE31448 datasets revealed that there was a positive correlation between protein SUMOylation and E2F-targets, MYC-targets-V1, Mtorc1-signaling, mitotic-spindle, G2M-checkpoint, and unfolded protein response (r>0.3, Figure 7A,B,C,D,E). In addition, a positive correlation was also observed between protein SUMOylation and mitotic-spindle, G2M-checkpoint, and unfolded protein response in the intersection of TCGA, GSE53752, GSE65216, and GSE31448 normal tissues datasets (r>0.3, Figure 7F,G,H,I,J). The intersection of these two arrays was shown in Figure 7K, with three overlapping pathways were found (mitotic-spindle, G2M-checkpoint, and unfolded protein response).
Next, we analyzed the pathway activity of E2F-targets, MYC-targets-V1, Mtorc1-signaling, mitotic-spindle, G2M-checkpoint, and unfolded protein response in the TCGA and GEO databases. These six pathways were up-regulated in TNBC tissues compared with adjacent normal breast tissues in TCGA, GSE53752, GSE65216, and GSE31448 databases (Figure 8A,B,C,D). Finally, meta-analysis revealed that the pathway activity of E2F-targets, MYC-targets-V1, Mtorc1-signaling, mitotic-spindle, G2M-checkpoint, and unfolded protein response were increased in TNBC (Figure 8E,F,G,J).
Discussion
Triple-negative breast cancer is characterized by high invasiveness and has a worse prognosis compared to other subtypes of breast cancer. Given the lack of ER, PR, and HER2 expression, there is no specific systemic treatment, such as endocrine therapy or anti-HER2 targeted therapy. Currently, the basis of TNBC treatment is surgery, chemotherapy, and radiotherapy. An anthracycline- and taxane-based chemotherapy regimen is the standard treatment for the prevention of TNBC recurrence and survival improvement (5). Early breast cancer trialists collaborative group (EBCTCG) analysis demonstrated a moderate reduction in 5- and 10-year risk of recurrence and death with dose intensity adjuvant chemotherapy, especially for TNBC patients (6). Yet, some patients still develop a rapid onset of recurrence and poor prognosis after conventional chemotherapy. Thus, identification of novel biomarkers that could be used to predict chemotherapy response and promising therapeutic targets might be beneficial in the treatment of TNBC.
Previous studies have indicated that SUMOylation is closely related to carcinogenesis, tumor proliferation, and metastasis, and is significantly up-regulated in most cancers (21-24). Therefore, SUMOylation may become a potential target for cancer treatment. However, the expression and underlying mechanisms of SUMOylation remain poorly understood in TNBC. In the present study, we advanced the knowledge regarding the role of SUMOylation in TNBC. We demonstrated that the pathway activity of protein SUMOlyation and the mRNA expression of SUMO1/2/3 were increased in TNBC tissues compared with adjacent normal breast tissues in TCGA and GEO databases. Meanwhile, our IHC staining results suggested that the expression of SUMO1/2/3 proteins was significantly increased in the tumor tissues of 211 TNBC patients. According to the survival analysis, SUMO1/2/3 protein expression levels were associated with the DFS and OS of TNBC patients. In addition, we developed a novel prognostic tool based on the IHC scores of SUMO1/2/3 to improve the prediction of disease recurrence for TNBC patients. Further use of the SB classifier might allow for better identification of TNBC patients who are most likely to benefit from chemotherapy. Therefore, the classifier for TNBC patients is both a prognostic and predictive tool. Patients with a SB classifier-defined low score might have both a lower likelihood of recurrence and a clear benefit from chemotherapy.
Moreover, we analyzed the pathways associated with the SUMOylation in TNBC. Our data showed that E2F-targets, MYC-targets-V1, Mtorc1-signaling, mitotic-spindle, G2M-checkpoint, and unfolded protein response were positively correlated with SUMOylation in the tumor tissues of TNBC patients. However, only mitotic-spindle, G2M-checkpoint, and unfolded protein response were identified to be positively correlated with SUMOylation in the normal tissues of TNBC patients. MYC is an important transcription factor. MYC mutations lead to uncontrolled expression of many genes, some of which are involved in cell proliferation and relate to the development of cancer. The MYC protein activates the transcription of SUMO activating enzyme subunit 1 (SAE1) by directly binding to the classic E-Box sequence located near the SAE1 transcription start site (25). Inhibition of SUMOylation was reported to disable MYC-induced cell proliferation and trigger G2/M cell cycle arrest in mouse and human MYC-driven lymphomas (26). In addition, there is accumulating evidence that SUMO directly and indirectly regulates protein localization within the mitotic spindle. AMP-activated protein kinase (AMPK) inhibits protein synthesis through suppression of mammalian target of rapamycin complex 1 (mTORC1). SUMOylation of AMPKα1 attenuates AMPK activation, and subsequently prompts the restoration of mTORC1 signaling (27). Retinoblastoma protein (Rb) is a prototypical tumor suppressor; hypo-phosphorylation of Rb is related to G0/G1 arrest by inhibiting the activity of E2F transcription factors, while hyper-phosphorylation of Rb releases E2F and converts the cell cycle from G0/G1 into S phase. SUMOylation of Rb causes Rb hyper-phosphorylation and E2F-1 release (28). X-box binding protein 1 (XBP1) is a key transcription factor that regulates the endoplasmic reticulum stress response, which is a cytoprotective mechanism that deals with the accumulation of unfolded protein in the endoplasmic reticulum. When endoplasmic reticulum stress occurs, unspliced XBP1 mRNA is converted into mature mRNA, and the transcription factor pXBP1 is translated. The transcription of endoplasmic reticulum-related genes is also activated to process unfolded proteins (29). SUMO-conjugase and UBC9 specifically bind to the leucine zipper motif of pXBP1 and increase the stability of pXBP1. Our analysis provides insights regarding the possible mechanism that the activation of SUMOylation was induced by MYC signaling, which eventually results in the activation of E2F-targets, Mtorc1-signaling, mitotic-spindle, G2M-checkpoint, and unfolded protein response.
The major strengths of the present study are that it had a large enough sample size of TNBC patients to perform survival analysis based on SUMO1/2/3 proteins, and developed a prognostic nomogram. In addition, some small molecule drugs that inhibit SUMOylation have been considered for the treatment of cancer. SUMO E1 inhibitor ML-792 is currently being tested in a phase 1 clinical trial for patients with metastatic solid tumors and lymphomas. In the current era of precision medicine, using a prognostic biomarker to select eligible patients and administration of specific treatments is a promising strategy. Our findings suggested that the inhibition of SUMOylation could be a promising therapeutic strategy for the treatment of TNBC patients.
Undoubtedly, there were several limitations in this study. Firstly, all TNBC patients were Chinese and from a single center, and thus, the findings of the present study may not be generalizable to all populations. Secondly, the sample size for this study was still limited to establish an external validation cohort and perform stratified analyses for specific subtypes of TNBC. Lastly, more intensive studies are still warranted to illustrate the underlying mechanisms in regulation of SUMOylation for TNBC.
Conclusions
In summary, our findings indicated that pathway activity of SUMOylation, as well as SUMO1/2/3 mRNA and protein levels were up-regulated in triple-negative breast cancer patients based on TCGA, GEO, and 212 TNBC specimens. The three SUMOs-based prognostic model could effectively classify TNBC patients into groups at a low- and high-risk of disease recurrence. Moreover, our study demonstrated that the SB classifier might be a useful predictive tool for TNBC patients treated with chemotherapy (Figure 9). Thus, the SB classifier potentially offers clinical value in directing personalized therapeutic regimen selection for TNBC patients. Furthermore, our analysis provides insights regarding the possible mechanism that the activation of SUMOylation was induced by MYC signaling, which ultimately results in the activation of E2F-targets, Mtorc1-signaling, mitotic-spindle, G2M-checkpoint, and unfolded protein response.
Acknowledgments
Funding: This research was funded by the Natural Science Foundation of Fujian Province (2017J01200 and 2020J01311402), the joint funds for the Innovation of Science and Technology of Fujian Province (2019Y9054), National Natural Science Foundation of China (81672817), National Science Foundation for Young Scientists of China (82003095), and the Fujian Provincial Key Laboratory of Hepatic Drug Research (KFLX2020001).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/gs-21-37
Data Sharing Statement: Available at http://dx.doi.org/10.21037/gs-21-37
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/gs-21-37). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Research Ethics Committee of Fujian Medical University Union Hospital (No. 2019KJCX011). Informed consent was obtained from each participant. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dent R, Trudeau M, Pritchard KI, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 2007;13:4429-34. [Crossref] [PubMed]
- Burstein MD, Tsimelzon A, Poage GM, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res 2015;21:1688-98. [Crossref] [PubMed]
- Abramson VG, Lehmann BD, Ballinger TJ, et al. Subtyping of triple-negative breast cancer: implications for therapy. Cancer 2015;121:8-16. [Crossref] [PubMed]
- Denkert C, Liedtke C, Tutt A, et al. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet 2017;389:2430-42. [Crossref] [PubMed]
- Budd GT, Barlow WE, Moore HC, et al. SWOG S0221: a phase III trial comparing chemotherapy schedules in high-risk early-stage breast cancer. J Clin Oncol 2015;33:58-64. [Crossref] [PubMed]
- Early Breast Cancer Trialists' Collaborative Group (EBCTCG). Increasing the dose intensity of chemotherapy by more frequent administration or sequential scheduling: a patient-level meta-analysis of 37 298 women with early breast cancer in 26 randomised trials. Lancet 2019;393:1440-52. [Crossref] [PubMed]
- Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet 2014;384:164-72. [Crossref] [PubMed]
- Liedtke C, Mazouni C, Hess KR, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol 2008;26:1275-81. [Crossref] [PubMed]
- Fornier M, Fumoleau P. The paradox of triple negative breast cancer: novel approaches to treatment. Breast J 2012;18:41-51. [Crossref] [PubMed]
- Wang Q, Zhang X, Chen L, et al. Regulation of the Expression of DAPK1 by SUMO Pathway. Biomolecules 2019;9:151. [Crossref] [PubMed]
- Satpathy S, Guerillon C, Kim TS, et al. SUMOylation of the ING1b tumor suppressor regulates gene transcription. Carcinogenesis 2014;35:2214-23. [Crossref] [PubMed]
- Dalmasso G, Nguyen HTT, Fais T, et al. Crohn's Disease-Associated Adherent-Invasive Escherichia coli Manipulate Host Autophagy by Impairing SUMOylation. Cells 2019;8:35. [Crossref] [PubMed]
- Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 2010;11:861-71. [Crossref] [PubMed]
- Wang QE, Zhu Q, Wani G, et al. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res 2005;33:4023-34. [Crossref] [PubMed]
- Becker J, Barysch SV, Karaca S, et al. Detecting endogenous SUMO targets in mammalian cells and tissues. Nat Struct Mol Biol 2013;20:525-31. [Crossref] [PubMed]
- Boulanger M, Paolillo R, Piechaczyk M, et al. The SUMO Pathway in Hematomalignancies and Their Response to Therapies. Int J Mol Sci 2019;20:3895. [Crossref] [PubMed]
- Chanda A, Chan A, Deng L, et al. Identification of the SUMO E3 ligase PIAS1 as a potential survival biomarker in breast cancer. PLoS One 2017;12:e0177639 [Crossref] [PubMed]
- Wang Q, Ye Y, Lin R, et al. Analysis of the expression, function, prognosis and co-expression genes of DDX20 in gastric cancer. Comput Struct Biotechnol J 2020;18:2453-62. [Crossref] [PubMed]
- Wang QS, Li F, Liao ZQ, et al. Low level of Cyclin-D1 correlates with worse prognosis of clear cell renal cell carcinoma patients. Cancer Med 2019;8:4100-9. [Crossref] [PubMed]
- Li F, Wang Q, Xiong X, et al. Expression of 4E-BP1 and phospho-4E-BP1 correlates with the prognosis of patients with clear cell renal carcinoma. Cancer Manag Res 2018;10:1553-63. [Crossref] [PubMed]
- Hendriks IA, Vertegaal AC. A comprehensive compilation of SUMO proteomics. Nat Rev Mol Cell Biol 2016;17:581-95. [Crossref] [PubMed]
- Bogachek MV, Park JM, De Andrade JP, et al. Inhibiting the SUMO Pathway Represses the Cancer Stem Cell Population in Breast and Colorectal Carcinomas. Stem Cell Reports 2016;7:1140-51. [Crossref] [PubMed]
- He X, Riceberg J, Soucy T, et al. Probing the roles of SUMOylation in cancer cell biology by using a selective SAE inhibitor. Nat Chem Biol 2017;13:1164-71. [Crossref] [PubMed]
- Li R, Wei J, Jiang C, et al. Akt SUMOylation regulates cell proliferation and tumorigenesis. Cancer Res 2013;73:5742-53. [Crossref] [PubMed]
- Amente S, Lavadera ML, Palo GD, et al. SUMO-activating SAE1 transcription is positively regulated by Myc. Am J Cancer Res 2012;2:330-4. [PubMed]
- Hoellein A, Fallahi M, Schoeffmann S, et al. Myc-induced SUMOylation is a therapeutic vulnerability for B-cell lymphoma. Blood 2014;124:2081-90. [Crossref] [PubMed]
- Yan Y, Ollila S, Wong IPL, et al. SUMOylation of AMPKalpha1 by PIAS4 specifically regulates mTORC1 signalling. Nat Commun 2015;6:8979. [Crossref] [PubMed]
- Meng F, Qian J, Yue H, et al. SUMOylation of Rb enhances its binding with CDK2 and phosphorylation at early G1 phase. Cell Cycle 2016;15:1724-32. [Crossref] [PubMed]
- Uemura A, Taniguchi M, Matsuo Y, et al. UBC9 regulates the stability of XBP1, a key transcription factor controlling the ER stress response. Cell Struct Funct 2013;38:67-79. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


