
The immune-related gene CD52 is a favorable biomarker for breast cancer prognosis
Introduction
Breast cancer is the most common type of carcinoma among females, a female that living in the United States with a 12.3% risk of suffering from breast cancer (1). With advances in diagnostic and therapeutic methods, the life expectancy of breast cancer patients can be increased. A proportion of breast carcinoma patients experience recurrence or progression of disease even after secretion, radiotherapy or chemotherapy treatment. Breast cancer remains the main cause of cancer-related deaths (2). Therefore, it is imperative to elucidate the underlying mechanisms of breast cancer progression, recurrence, and resistance.
To date, numerous reports have demonstrated that the tumor microenvironment and immune cell infiltrates play key roles in tumorigenesis and the development of breast carcinoma, and the tumor microenvironment not only contains the cancerous cells but also contains noncancer cells such as stromal and immune cells that may serve as important regulators for multiple cancer types (3-5). The different components of the tumor microenvironment may lead to different clinical outcomes for the cancer patient (6). In breast cancer patients, the immune system may have a dual role in disease progression and immune surveillance. With improved understanding of the association between the immune system and cancer biology, researchers have become increasingly aware of the importance of patient immunity in breast malignancies. Indeed, previous studies have detected the presence of significant immune-related genes in breast cancer patients (7).
Various immune cell types may have different functions in breast cancer. Tumor-associated neutrophils have been shown to act as immunosuppressors in breast cancer mouse models (8). Tumor-associated macrophages (TAMs) can be divided into M1 and M2 macrophages based on the biological processes (BP) involved. While M1 macrophages have been shown to have an anticancer role (9), M2 macrophages appear to promoting cancer growth and metastasis (10). Other immune cells such as tumor-infiltrating lymphocytes also play a key role in breast cancer (11). Several immune-related genes may affect tumor progression via regulating immune cell infiltrates into the tumor microenvironment. NanoString Immunology Gene Expression Panel revealed that tumor necrosis factor receptor-associated factor 1 (TRAF1), nuclear factor kappa B subunit 1 (NFKB1), and chemokine ligand 13 (CXCL13) were significantly related to tumor infiltrating lymphocyte levels and were good independent prognostic biomarkers for triple-negative breast cancer (12). Moreover, studies have shown a correlation between the expression of immune genes and response to trastuzumab therapy in human epidermal growth factor receptor 2 positive (HER2+) breast cancer patients (12).
This study investigated the significant immune-related genes for breast cancer. The study involved a discovery phase and two validation phases. In the discovery phase, the immune-related genes that showed a significant relationship with the prognosis of breast cancer were screened via bioinformatics analysis. In the first validation phase, the genes identified to be correlated with good prognosis were further evaluated using The Cancer Genome Atlas (TCGA) and a cohort based on a tissue chip. Compared to Wang et al. study (13), we not only assessed the immune protein CD52 in terms of its expression levels, effects on immune cell infiltrates, prognostic value, relationship with key immune-related genes, but also explored effects on the clinical status of the breast carcinoma such as the tumor type, lymph node, and metastasis in the second validation phase. To date, the CD52 is known as a cell surface glycoprotein that is a unknown function, it expresses in various immune cells such as lymphocytes, natural killer (NK) cells, monocytes, macrophages, and so on (14,15). The CD52 regularly was detectable in hematological malignancies such as T peripheral cell lymphoma (PTCL), cutaneous T cell lymphoma, and T cell prolymphocytic leukemia (16-20). However, the role of CD52 in solid tumor such as BRCA still unclear.
We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/gs-20-922).
Methods
Statistical analysis
The immune-stromal component in the tumor microenvironment (TME) was investigated using the R package ‘ESTIMATE’ (https://www.r-project.org/); The Kaplan-Meier method was used to perform the survival curves, and log-rank P<0.05 was considered statistically significant. The differential genes with a log fold change |logFC| >1 and false discovery rate FDR <0.05 were deemed as significant; The cluterProfiler packages and enrichplot packages were using the hypergeometric distribution to calculate each term of GO’s or KEGG’s P value and q-value. A P value and q-value less than 0.05 were considered as significant enrichment functions; The PPI network of differential genes is constructed through STRING website, and we selected the interacting proteins with a confidence score more than 0.7; The R package CIBERSORT was used to estimate the tumor infiltrating immune cells. P value <0.05 was seem as the significant difference.
Extraction of raw data
The mRNA-seq data and clinical information of 1,109 breast cancer patients and 113 healthy subjects were collated from the TCGA public database (https://portal.gdc.cancer.gov/).
Analyzing the immune, stromal, and estimate scores
The immune-stromal component in the tumor microenvironment (TME) was investigated using the package in the R language version 4.0.3 (https://www.r-project.org/). The results of this analysis showed three types of scores that included an immune score, stromal score, and an estimate score. The higher the respective score, the larger the proportion of the corresponding component in the TME.
Survival analysis
For survival analysis, the R language loaded with the survival and survminer package was used. Furthermore, the univariate Cox regression analyses and the Kaplan-Meier method were applied to assess the prognostic role of differentially expressed genes (DEGs). The Kaplan-Meier method was used to perform the survival curves, and log-rank P<0.05 was considered statistically significant.
Screening of the DEGs based on immune and stromal scores
The tumor samples were divided into two groups (high or low score groups) based on the median values of the immune and stromal scores. The gene differentiation analysis was conducted via Package “limma” and the DEGs were selected through comparison between the high and low score groups. The differential genes with a log fold change |logFC| >1 and false discovery rate FDR <0.05 were deemed as significant.
Enrichment function analysis
The potential biological functions and pathways of the DEGs in breast cancer were explored using the R language with the aid of the clusterProfiler, enrichplot, and ggplot2 packages. The cluterProfiler packages and enrichplot packages were using the hypergeometric distribution to calculate each term of GO’s or KEGG’s P value and q-value. A P value and q-value less than 0.05 were considered as significant enrichment functions.
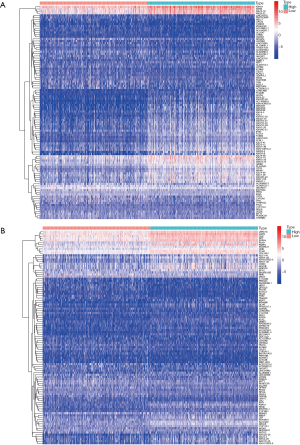
Construction of heatmaps
The R language with the pheatmap package was used to construct the heatmaps of the DEGs.
Analysis of the association between clinical characteristics and the immune-stromal scores
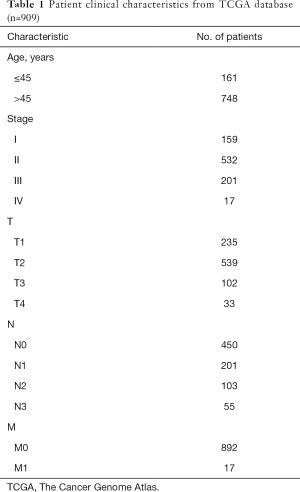
The clinical features data of the breast cancer patients were collated from the TCGA. Only 909 breast cancer cases showed detailed clinical characteristics. The analysis of the relationship between clinical characteristics and immune-stromal scores was conducted using R language. Wilcoxon rank sum or Kruskal-Wallis rank sum test was used to determine significance, and P<0.05 was considered statistically significant.
Construction of the protein-protein interaction (PPI) network
The PPI network of differential genes is constructed through string website, and the top 30 genes of degree are calculated by cytoHubba plug-in in Cytoscape software.
Tumor-infiltrating immune cell (TIC) profile
The CIBERSORT deconvolution algorithm was a method for characterizing cell composition of complex tissues from their gene expression profiles to estimate the relative abundance of 22 immune cells types. There were 845 cancer samples with P value <0.05 and these were applied to the final analysis.
Ethical Statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
The process of analysis of this study
All of these methods that workflow diagram of present study showing Figure 1.
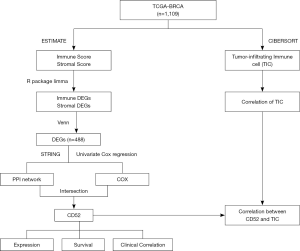
Results
The relationship between immune and stromal scores, and clinical features
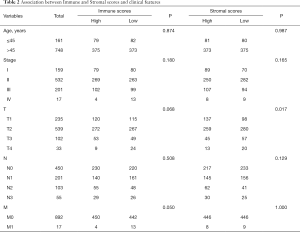
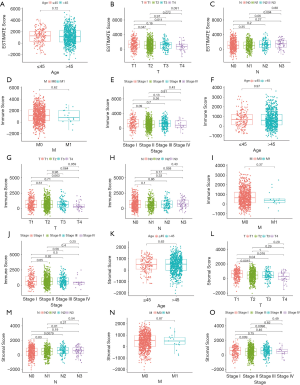
The immune scores, stomal scores, and clinical characteristics from breast cancer patients were extracted from the TCGA cohort (Table 1). The breast cancer patients were divided into two groups (high and low score groups) based on the median levels of the immune and stromal scores (Table 2). The relationship between immune and stromal scores, and breast cancer prognosis based on the TCGA cohort was investigated. The results demonstrated that Estimatescore did not have significantly influence on prognosis (Figure 2A, P=0.426), higher immune scores were correlated with better prognosis (Figure 2B, P=0.011). However, stromal scores did not have a significant role in the prognosis of breast cancer patients (Figure 2C, P=0.666). We also investigated the relationship between clinical features and immune and stromal scores. The results showed that age did not significantly influence immune scores. Patients with T1 stage cancer showed higher immune scores compared to patients with T2 (P=0.047) and T4 (P=0.011) stage cancers. Higher immune scores were detected in early stages of breast cancer, but there were no significant differences between early stage and advance stage cancer. The stromal scores were not significantly affected by age. Higher stromal scores were observed in early stage cancer and T1 staging showed the highest stromal scores. Stromal scores for T1 stage patients were significantly higher than that observed in T2 (P=0.0015), T3 (P=0.04), and T4 (P=0.016) patients. However, the stromal scores in lymph node metastasis were inconsistent with primary tumor size, distant metastasis, and the stages of breast cancer. The relationship between stromal scores and lymph node metastasis showed that compared to N0 and N1, N2 and N3 had higher stromal scores. There were significant differences between N3 and N1 (P=0.01), and N3 and N0 (P=0.0079). Stage I demonstrated significantly higher stromal scores compared to stage II (P=0.009), and stage III had a significantly higher stromal score compared to stage II (P=0.0096) (Figure 3).
Full table
Full table

Screening the differentially expressed genes (DEGs) associated with immune scores and stromal scores in breast cancer
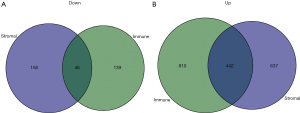
The TCGA cohort patients were divided into two groups consisting of high and low immune and stromal scores. The gene expression profiles between the high and low groups were examined. A total of 1,252 upregulated genes and 185 downregulated genes were identified based on the immune scores (high vs. low) (Figure 4). Based on the stromal scores (high vs. low), 1,079 upregulated genes and 204 downregulated genes were identified (Figure 4). A Venn analysis diagram was constructed to select the genes that were significantly related to stromal and immune cells. A total of 442 upregulated genes and 46 downregulated genes were found in the intersection (Figure 5). Herein, these genes were further analyzed for their function and prognostic role in breast cancer.
Enrichment functions of the DEGs in the Venn diagram intersection
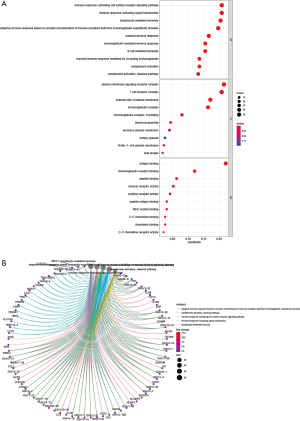
The potential functions of the DEGs identified in the Venn diagram were further investigated using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. The enrichment functions analyses revealed the top five BP including adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains, lymphocyte mediated immunity, immunoglobulin mediated immune response, immune response-activating signal transduction, and immune response-activating cell surface receptor signaling pathway. The top five cellular components (CC) included plasma membrane signaling receptor complex, immunoglobulin complex, circulating external side of plasma membrane, blood microparticle, and immunoglobulin complex. The molecular functions (MF) identified included antigen binding, immunoglobulin receptor binding, immune receptor activity, cytokine receptor activity, and major histocompatibility complex (MHC) protein binding (Figure 6). The KEGG pathway analysis revealed that the DEGs function as the part of the viral protein interaction with cytokine-cytokine receptors, hematopoietic cell lineage, malaria, interaction between cytokines and cytokine receptors, and cell adhesion molecules (Figure 7). All these results suggested that the screened DEGs have essential roles in the immune system.

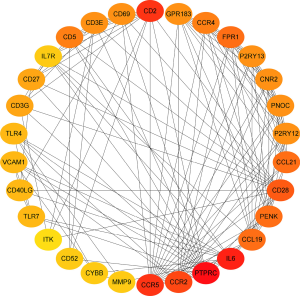
Construction of the PPI network for the screened DEGs
To investigate the relationship between the screened top 30 DEGs, the PPI network was constructed (Figure 8).
The prognostic role of the DEGs in breast carcinoma
The top 30 genes which interacted with the DEGs and included CD52 (Figure 9A). Univariate Cox regression analyses were conducted to explore the prognostic value of the selected DEGs. The results showed that 5 genes predicted favorable or adverse prognosis for breast cancer (P<0.05, Figure 9B). Venn analysis was used to further screen the top 30 interacting genes that significantly affected prognosis (Figure 9C) and CD52 was detected in the intersection. This further confirmed that CD52 may have an essential role in the prognosis and immune response in breast cancer patients.

The expression and prognostic value of CD52 in breast cancer
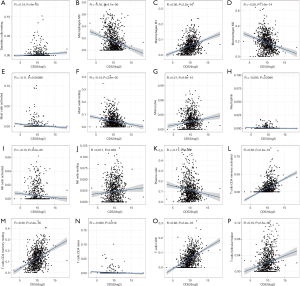
The transcriptional levels of CD52 in breast cancer patients and healthy patients were investigated using the TCGA cohort. Tumor samples showed significant overexpression of CD52 (P=0.033) compared to healthy samples (Figure 9D,E). Univariate analysis indicated that overexpression of CD52 may be an effective favorable biomarker for breast cancer patients (Figure 9F). The relationship between CD52 expression levels and cancer stages showed in Table 3. Further, The subgroup analysis of CD52 expression levels based on age and the status of the malignancy revealed that the early stages of disease were associated with higher CD52 expression levels compared to advanced stages (Figure 9G,H,I,J,K). These results indicated that CD52 expression level may predict the stages of breast malignancies.
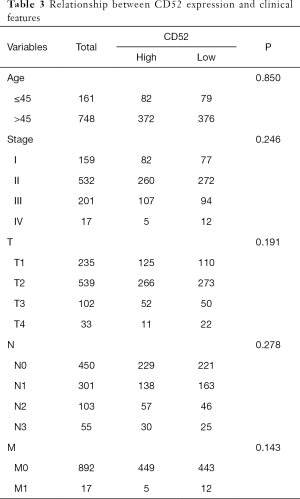
Full table
Analysis of the immune infiltrates in breast cancer patients
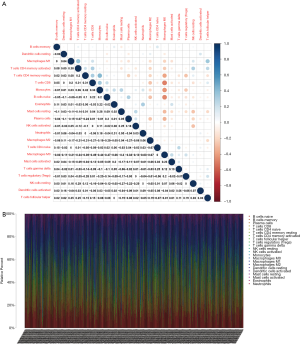
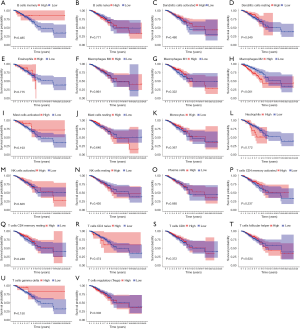
The levels of immune infiltrates and the relationship between different cellular immune infiltrates were examined using the TCGA breast cancer cohort (Figure 10A,B). The results demonstrated that M0 macrophages may be the most important negative regulator for resting dendritic cells, M1 macrophages, activated memory CD4 T cells, resting memory CD4 T cells, CD8 T cells, monocytes, naïve B cells, eosinophils, resting mast cells, and plasma cells. However, CD8 T cells may be a positive regulator for memory B cells, resting dendritic cells, M1macrophages, activated memory CD4 T cells, resting memory CD4 T cells, and monocytes. Further, the breast cancer patients were divided into two group based on the mean CD52 expression. The relationship between CD52 expression levels and immune infiltrates was explored. Overexpression of CD52 led to higher numbers of M1 macrophages, monocytes, follicular helper T cells, and resting memory CD4 T cells. Downregulation of CD52 resulted in increased numbers of M2 macrophages, resting mast cells and plasma cells, and M0 macrophages. The association between CD52 and the different immune cell infiltrates is shown in Figure 11. Finally, the prognostic role of different immune infiltrates in breast cancer was investigated. The data showed that higher numbers of M2 macrophages led to poor prognosis (P<0.001), but the levels of the other immune cell infiltrates did not have a significant influence on breast cancer prognosis (Figure 12).
Discussion
This study included a discovery phase and three validation phases. In the discovery phase, CD52 was identified as a significant immune-related gene. In the validation phases, high levels of CD52 expression were related to different stages of breast malignancy, and the levels of CD52 expression may have positive or negative correlations with different immune cell infiltrates. Furthermore, high expression of CD52 may function as an effective favorable biomarker for breast cancer patients.
CD52 is one of most abundant membrane glycoproteins and is expressed on the surface of normal or malignant lymphocytes, monocytes, and macrophages. It is also expressed on epithelial cells and in the male reproductive tract (21). For lymphocytes, there are approximately 450,000 molecules covering about 5% of the cell surface. Considering this, CD52 may be an excellent therapeutic target for several cancer types. CD52 is usually observed in hematological malignancies. A study by Ginaldi et al. revealed that the differential expression of CD52 was detected in different populations of lymphoid leukemias, and the levels of expression may be used as a biomarker for responsiveness to therapy (22). Alemtuzumab, a monoclonal antibody that targets CD52, has been used in the clinical treatment of chronic lymphocytic leukemia (CLL) (23). The anti-cancer mechanisms of alemtuzumab in CLL are mediated by caspase-independent apoptosis (24). For CLL, resistance to fludarabine and rituximab has been associated with downregulation of CD20 and upregulation of CD52 (25), suggesting that the levels of CD52 expression may be associated with drug sensitivity. Alemtuzumab is also used for the treatment of peripheral T-cell lymphoma (PTCL) (26). All these reports demonstrate the essential role of CD52 in tumorigenesis, disease progression, and response to therapy. However, to date, CD52-related research in the cancer field has primarily focused on hematological malignancies, with few studies investigating the relationship between CD52 and solid tumors such as breast cancers. Furthermore, the functions of CD52 in cancer is yet to be fully elucidated.
The present study identified CD52 as a significant immune-related gene which may act as an effective biomarker for breast cancer. The investigations demonstrated that CD52 was positively correlated with various immune cells including T cell types and M1 macrophages, but CD52 was negatively correlated with infiltration of M2 macrophages. In breast cancer patients, elevated expression of immune-related genes have been associated with longer progression-free survival, even in patients with aggressive breast cancer subtypes such as HER2 positive malignancies (27). High expression of lymphocyte-associated genes in node-negative HER2 positive breast cancers have been correlated with lower recurrence rates (28). Studies have demonstrated that the T-cell metagene can predict a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers (29). Our results were consistent with these reports and demonstrated that higher expression of key immune-related genes were associated with improved prognosis for breast cancer. However, different immune cells may have different functions in cancer. Herein, the association between different immune cells and breast cancer prognosis was examined. Over the past decades, numerous studies have observed the infiltration of TAMs into tumor tissues. However, increasing evidence have indicated that TAMs promote multiple cancer types (3,30). In this current study, higher numbers of M2 macrophages corresponded to poorer breast cancer prognosis. Yamaguchi et al. demonstrated that the M2 macrophages can promote peritoneal gastric cancer (31) and studies by Chen and colleagues revealed that M2 macrophages enhanced cancer metastasis via production of chitinase 3-like protein 1 (CHI3L1) (32). TAMs not only directly influence cancer cells but can also affect cancer progression by inhibiting host anti-cancer immunity via several mechanisms (33,34). All these reports are consistent with our observations that higher levels of M2 macrophage infiltration led to poorer prognosis in cancer patients. The limitation of our study is that absence of validation by experiments.
Conclusions
CD52 is a significant immune-related gene and may be used as a favorable biomarker for breast cancer. The expression of CD52 may be effective in predicting the prognosis and stages of breast carcinoma, with high expression of CD52 indicative of early stages and improved prognosis. The potential functions of CD52 in the tumor microenvironment include inhibition of M2 macrophages infiltration, as well as the promotion of T cells, M1 macrophages, and plasma cells into the tumor microenvironment to exert anti-cancer activity. CD52 can act as the potential immune therapy for BRCA.
Acknowledgments
Funding: (I) General program of Guangxi Natural Science Foundation (No. 2019JJA140071). (II) The First Batch of High-level Talent Scientific Research Projects of the Affiliated Hospital of Youjiang Medical University for Nationalities in 2019 (Contract No. R20196307). (III) Project of Youjiang Medical University for Nationality (No. YY2019ky001). (IV) Project of National Natural Science Foundation (No. 81600541).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/gs-20-922
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/gs-20-922). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- DeSantis C, Ma J, Bryan L, et al. Breast cancer statistics, 2013. CA Cancer J Clin 2014;64:52-62. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Choi J, Gyamfi J, Jang H, et al. The role of tumor-associated macrophage in breast cancer biology. Histol Histopathol 2018;33:133-45. [PubMed]
- Houthuijzen JM, Jonkers J. Cancer-associated fibroblasts as key regulators of the breast cancer tumor microenvironment. Cancer Metastasis Rev 2018;37:577-97. [Crossref] [PubMed]
- Lim B, Woodward WA, Wang X, et al. Inflammatory breast cancer biology: the tumour microenvironment is key. Nat Rev Cancer 2018;18:485-99. [Crossref] [PubMed]
- Zhang C, Cheng W, Ren X, et al. Tumor Purity as an Underlying Key Factor in Glioma. Clin Cancer Res 2017;23:6279-91. [Crossref] [PubMed]
- Zhu B, Tse LA, Wang D, et al. Immune gene expression profiling reveals heterogeneity in luminal breast tumors. Breast Cancer Res 2019;21:147. [Crossref] [PubMed]
- Eruslanov EB, Bhojnagarwala PS, Quatromoni JG, et al. Tumor-associated neutrophils stimulate T cell responses in early-stage human lung cancer. J Clin Invest 2014;124:5466-80. [Crossref] [PubMed]
- Tang X. Tumor-associated macrophages as potential diagnostic and prognostic biomarkers in breast cancer. Cancer Lett 2013;332:3-10. [Crossref] [PubMed]
- Medrek C, Pontén F, Jirström K, et al. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer 2012;12:306. [Crossref] [PubMed]
- Mohammed ZM, Going JJ, Edwards J, et al. The relationship between lymphocyte subsets and clinico-pathological determinants of survival in patients with primary operable invasive ductal breast cancer. Br J Cancer 2013;109:1676-84. [Crossref] [PubMed]
- Lee HJ, Lee JJ, Song IH, et al. Prognostic and predictive value of NanoString-based immune-related gene signatures in a neoadjuvant setting of triple-negative breast cancer: relationship to tumor-infiltrating lymphocytes. Breast Cancer Res Treat 2015;151:619-27. [Crossref] [PubMed]
- Wang J, Zhang G, Sui Y, et al. CD52 Is a Prognostic Biomarker and Associated With Tumor Microenvironment in Breast Cancer. Front Genet 2020;11:578002. [Crossref] [PubMed]
- Hale G, Xia MQ, Tighe HP, et al. The CAMPATH-1 antigen (CDw52). Tissue Antigens 1990;35:118-27. [Crossref] [PubMed]
- Waldmann H, Hale G. CAMPATH: from concept to clinic. Philos Trans R Soc Lond B Biol Sci 2005;360:1707-11. [Crossref] [PubMed]
- Dearden CE, Matutes E, Cazin B, et al. High remission rate in T-cell prolymphocytic leukemia with CAMPATH-1H. Blood 2001;98:1721-6. [Crossref] [PubMed]
- Enblad G, Hagberg H, Erlanson M, et al. A pilot study of alemtuzumab (anti-CD52 monoclonal antibody) therapy for patients with relapsed or chemotherapy-refractory peripheral T-cell lymphomas. Blood 2004;103:2920-4. [Crossref] [PubMed]
- Lundin J, Hagberg H, Repp R, et al. Phase 2 study of alemtuzumab (anti-CD52 monoclonal antibody) in patients with advanced mycosis fungoides/Sezary syndrome. Blood 2003;101:4267-72. [Crossref] [PubMed]
- Pawson R, Dyer MJ, Barge R, et al. Treatment of T-cell prolymphocytic leukemia with human CD52 antibody. J Clin Oncol 1997;15:2667-72. [Crossref] [PubMed]
- Zhang Z, Zhang M, Goldman CK, et al. Effective therapy for a murine model of adult T-cell leukemia with the humanized anti-CD52 monoclonal antibody, Campath-1H. Cancer Res 2003;63:6453-7. [PubMed]
- Hale G, Rye PD, Warford A, et al. The glycosylphosphatidylinositol-anchored lymphocyte antigen CDw52 is associated with the epididymal maturation of human spermatozoa. J Reprod Immunol 1993;23:189-205. [Crossref] [PubMed]
- Ginaldi L, De Martinis M, Matutes E, et al. Levels of expression of CD52 in normal and leukemic B and T cells: correlation with in vivo therapeutic responses to Campath-1H. Leuk Res 1998;22:185-91. [Crossref] [PubMed]
- Ferrajoli A, O'Brien SM, Cortes JE, et al. Phase II study of alemtuzumab in chronic lymphoproliferative disorders. Cancer 2003;98:773-8. [Crossref] [PubMed]
- Nückel H, Frey UH, Röth A, et al. Alemtuzumab induces enhanced apoptosis in vitro in B-cells from patients with chronic lymphocytic leukemia by antibody-dependent cellular cytotoxicity. Eur J Pharmacol 2005;514:217-24. [Crossref] [PubMed]
- Ashraf U, Elefante AN, Cruz R, et al. Emergence of Rituximab-Fludarabine Resistance Is Associated with Changes in CD20 Antigen Expression and Improved Response to Almetuzumab Therapy in Patients with Chronic Lymphocytic Leukemia (CLL). Blood 2007;110:2054. [Crossref]
- Zinzani PL, Alinari L, Tani M, et al. Preliminary observations of a phase II study of reduced-dose alemtuzumab treatment in patients with pretreated T-cell lymphoma. Haematologica 2005;90:702-3. [PubMed]
- Matikas A, Lövrot J, Ramberg A, et al. Dynamic evaluation of the immune infiltrate and immune function genes as predictive markers for neoadjuvant chemotherapy in hormone receptor positive, HER2 negative breast cancer. Oncoimmunology 2018;7:e1466017. [Crossref] [PubMed]
- Alexe G, Dalgin GS, Scanfeld D, et al. High expression of lymphocyte-associated genes in node-negative HER2+ breast cancers correlates with lower recurrence rates. Cancer Res 2007;67:10669-76. [Crossref] [PubMed]
- Faucheux L, Grandclaudon M, Perrot-Dockès M, et al. A multivariate Th17 metagene for prognostic stratification in T cell non-inflamed triple negative breast cancer. Oncoimmunology 2019;8:e1624130. [Crossref] [PubMed]
- Komohara Y, Fujiwara Y, Ohnishi K, et al. Tumor-associated macrophages: Potential therapeutic targets for anti-cancer therapy. Adv Drug Deliv Rev 2016;99:180-5. [Crossref] [PubMed]
- Yamaguchi T, Fushida S, Yamamoto Y, et al. Tumor-associated macrophages of the M2 phenotype contribute to progression in gastric cancer with peritoneal dissemination. Gastric Cancer 2016;19:1052-65. [Crossref] [PubMed]
- Chen Y, Zhang S, Wang Q, et al. Tumor-recruited M2 macrophages promote gastric and breast cancer metastasis via M2 macrophage-secreted CHI3L1 protein. J Hematol Oncol 2017;10:36. [Crossref] [PubMed]
- Parker KH, Sinha P, Horn LA, et al. HMGB1 enhances immune suppression by facilitating the differentiation and suppressive activity of myeloid-derived suppressor cells. Cancer Res 2014;74:5723-33. [Crossref] [PubMed]
- Anderson KM, Czinn SJ, Redline RW, et al. Induction of CTLA-4-mediated anergy contributes to persistent colonization in the murine model of gastric Helicobacter pylori infection. J Immunol 2006;176:5306-13. [Crossref] [PubMed]


