
Whole-exon sequencing insights into pancreatic synovial sarcoma: a case report
Introduction
Pancreatic synovial sarcoma (PSS) is rare, and the genome description of this disease is seldom studied (1). More than 30 cases of synovial sarcoma (SS) are sequenced on all organs and uploaded in the cBioPortal database, while PSS has not yet been sourced (2). Therefore, we performed a whole exon DNA sequencing (a tumor sample and a normal sample) on a case of primary PSS. The function of the PSS in chromosome remodeling was found in the sequencing (arid1a, arid1b, tp53, etc.). The driver gene mutation and the type of mutation in this patient is not very consistent with that was reported on the cBioPortal (incidentally, the SS on the cBioPortal is mostly other tissues and organs other than the pancreas). In summary, we performed an exon analysis of the PSS genome and found its role in the chromosome remodeling, and to increase understanding of the pathogenesis in this case to widen the therapeutic possibilities.
We present the following case in accordance with the CARE reporting checklist (available at http://dx.doi.org/10.21037/gs-20-680).
Case presentation
A 66-year-old male with a previous history of oral cancer was hospitalized because of an upper abdominal pain for half a month. His oral cancer located in the tongue root and tonsil without systemic metastasis was treated by extended radical resection followed by radiotherapy and chemotherapy (paclitaxel and nedaplatin). The histopathological diagnosis of his oral cancer was high-grade differentiated squamous cell carcinoma in the lingual root and tonsil with striated muscle invasion and cancer metastasis in the lymph node.
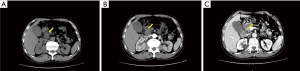
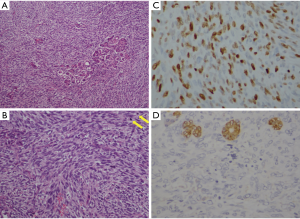
After 19 months of the surgery, he came to the hospital complaining about a sudden upper abdominal pain. Upper abdomen enhanced computed tomography (CT) documented an occupying position and was inclined to consider malignant tumor in the pancreatic head, which has invaded the lower part of the common bile duct, with biliary dilatation and mild expansion of the pancreatic duct (Figure 1). Further studies including blood biochemical examination and tumor markers showed the abnormal elevation in the alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), glutamyltranspeptidase (GLT) and total bilirubin (TB), direct bilirubin (DB), indirect bilirubin (IB) but without abnormalities in the tumor markers. Therefore, we performed laparoscopic exploration after the open pancreaticoduodenectomy. During the intraoperative seeing, the liver cholecystitis was thickened gallbladder wall, the common bile duct was widened, the gallbladder was swollen, the pancreas was medium in texture, and the head of the pancreas was 3 cm in size, which was close to the portal vein and the cut surface was gray. Apart from multiple lymph nodes in the hepatoduodenal ligament, there were no obvious abnormalities were found in the remaining abdominal cavity. Postoperative pathological results showed that the size of the spindle cell malignancy was 2.5 cm × 2.5 cm and the histopathological diagnosis was PSS (Figure 2A,B). Immunohistochemical result (Figure 2C,D): EMA (−), CD34 (−), CD117 (−), S-100 (−), Desmin (−), SMA (−), DOG1 (−), Ki-67 (+50%), CK-pan (−), Bcl-2 (+), CD99 (−). No adjuvant treatment was administered. The patient was alive and well at two years.
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient. This study was ethically approved by Ethics Committee of Sir Run Shaw Hospital of Zhejiang University (No. 20160222-9).
Whole-exon sequencing
After obtaining the right to use the patient’s specimen, we conducted whole exome sequencing of the tumor and paired paratumorous tissue to explore the possible molecular alterations (Lian-Chuan Biological Technology Co., Ltd, Hangzhou, China). DNA was extracted from the tissue samples using a DNeasy Tissue Kit and was subjected to Whole Exome Sequencing. After being quantified by the Qubit 3.0 Fluorometer (Thermo scientific, MA, USA), DNA was mechanically fragmented on an E220 focused ultrasonicator Covaris (Covaris, Woburn, MA, USA). xGen Exome Research Panel (Integrated DNA Technologies) was applied to prepare the DNA library and exome capture. When the quality control was passed and quantification was finished by Agilent 2100 Bioanalyzer (Agilent Technologies) and Qubit 3.0 Fluorometer, sequencing was begun using Illumina PE150 (lllumina Inc., San Diego, CA, USA).
Firstly, we found that there are many copy number variations (CNV) of exon gene in all the 24 chromosomes. Of them, chr1, chr2, chr4 have the most exon gene amplifications and chr21, chr22, chrY have the most exon gene deletions (Figure 3). Then we performed GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis to identify the potential molecular function of these driver-genes. GO analysis showed that many driver-genes are related to chromosome or chromatin organization, chromatin modification and its regulation process (Figure 4A). KEGG analysis demonstrated that many driver-genes are enriched in the cancer-related pathways, such as “Pathways in cancer”, “MicroRNA in cancer”, “Transcriptional mis-regulation in cancer” and so on (Figure 4B).

Besides, we found mutations or CNV of five vital driver-genes in the results of sequencing, including arid1a, arid1b, tp53, cdkn2a and asxl1 (Table 1). Of them, arid1a and arid1b in exon 1 both are in-frame mutations, but the mutations are not on the functional area. However, tp53 is a hotspot mutation in the encoding sequence c.524G>A (p.Arg175His) in exon 5 with a single nucleotide variation (SNV). Cdkn2a is a frameshift mutation at the encoding sequence c.169_184del (p.Ala57CysfsTer84) in exon 2 and asxl1 is another frameshift mutation at the encoding sequence c.1934del (p.Gly645ValfsTer58) in exon 5, which may be related to the incidence of SS (Figure 5).
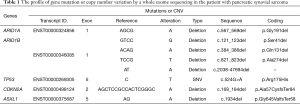
Full table
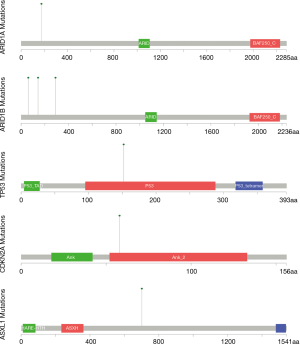
Discussion
SS derived from mutation of mesenchymal cells is a malignant tumor with the characteristic of frequent recurrence within two years and prone to metastasis (3). Intra-abdominal SS commonly occurs in the retroperitoneum and typically is presented with abdominal pain or compression of adjacent organs (4). The standard treatment of SS is to expand the resection area with negative margin followed by radiation and/or chemotherapy. The overall 5-year survival is from 36 to 76%. Age, tumor size, cell differentiation and type of gene translocation are predictive prognostic factors for the patients with SS (5).
Owing to the features of metastases, there have been several case reports about the pancreatic metastasis from SS. For example, Makino et al. (6) reported a case report of pancreatic metastasis from SS was successfully treated by metastasectomy with adjuvant chemotherapy. Yamamoto et al. (7) reported a case of surgical treatment for pancreatic metastasis from SS in the right thigh.
However, the primary SS is very rare and the related study has not been exploited. Only Luc et al. (8) previously reported a case of primary monophasic SS of the pancreas in a 44-year-old male who started with multiple episodes of retroperitoneal hemorrhage and underwent distal pancreatectomy without complication and adjuvant treatment. The patient is alive at 2 years. In our case, the 66-year-old patient came to the hospital for the upper abdominal pain and underwent a laparoscopic exploration followed by open pancreaticoduodenectomy without adjuvant treatment and the patient was also alive and well at 2 years, which indicates that the surgical intervention is an effective approach under this condition.
Because the incidence of SS is low, the genetic changes associated with its pathogenesis are rarely studied. In the current study, we tried to identify its cause in terms of gene variations by the use of whole exome sequencing and found that this case had mutations in the chromosomal remodeling and chromosome modification of the driving genes through the GO and KEGG analysis. We then focused on the five important driver-genes, namely, arid1a, arid1b, tp53, cdkn2a and asxl1. Of them, tp53 is a hotspot mutation, cdkn2a and asxl1 are frameshift mutations, which may be related to the pathogenesis of SS. Comparing with the results about SS in the cBioPortal, hotspot mutation of tp53 and frameshift mutation of cdkn2a were also found in the database based on analysis in more than 10 samples of SS. For example, Subramaniam et al. (9) reported the p16INK4A (cdkn2a) gene deletion is a frequent genetic event in SSs. Sabah et al. (10) also found that loss of p16INK4A expression is associated with allelic imbalance/loss of heterozygosity of chromosome 9p21 in microdissected SSs. However, the asxl1 frameshift mutation was firstly found by our team, which may be particularly relevant with the primary PSS and potentially related treatment target.
In conclusion, we reported a whole-exon sequencing insight into PSS and found some vital gene mutations, such as tp53, cdkn2a, asxl1, which may be related with the occurrence and targeted treatment of primary PSS. However, due to the limited samples of PSS, the results could be biased and larger samples are needed to validate these findings. In addition, a follow-up study is required to verify our findings. Conceivably, therapeutic strategies proposed here need further preclinical and clinical studies. In this study, WES is beneficial to providing evidence for the diagnosis of rare disease and five important mutations in genes were identified. These findings provided treatment guidance for this patient and a new idea for the diagnosis of PSS. Meanwhile, higher sequencing depth and integrated omics data, like epigenomics, transcriptomics and proteomics, may help to generate a fine structure.
Acknowledgments
Funding: This work was supported by the Zhejiang Clinical Research Center of Minimally Invasive Diagnosis and Treatment of Abdominal Diseases (Grant No. 2018E50003) and Educational Commission of Zhejiang Province of China (Grant No. Y201738035).
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at http://dx.doi.org/10.21037/gs-20-680
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/gs-20-680). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was ethically approved by Ethics Committee of Sir Run Shaw Hospital of Zhejiang University (No. 20160222-9). All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferrari A, De Salvo GL, Brennan B, et al. Synovial sarcoma in children and adolescents: the European Pediatric Soft Tissue Sarcoma Study Group prospective trial (EpSSG NRSTS 2005). Ann Oncol 2015;26:567-72. [Crossref] [PubMed]
- Nielsen TO, Poulin NM, Ladanyi M. Synovial sarcoma: recent discoveries as a roadmap to new avenues for therapy. Cancer Discov 2015;5:124-34. [Crossref] [PubMed]
- El Beaino M, Araujo DM, Lazar AJ, et al. Synovial Sarcoma: Advances in Diagnosis and Treatment Identification of New Biologic Targets to Improve Multimodal Therapy. Ann Surg Oncol 2017;24:2145-54. [Crossref] [PubMed]
- Fisher C, Folpe AL, Hashimoto H, et al. Intra-abdominal synovial sarcoma: a clinicopathological study. Histopathology 2004;45:245-53. [Crossref] [PubMed]
- Trassard M, Le Doussal V, Hacène K, et al. Prognostic factors in localized primary synovial sarcoma: a multicenter study of 128 adult patients. J Clin Oncol 2001;19:525-34. [Crossref] [PubMed]
- Makino Y, Shigekawa M, Kegasawa T, et al. A case report of pancreatic metastasis from synovial sarcoma successfully treated by metastasectomy with adjuvant chemotherapy. Medicine (Baltimore) 2016;95:e4789 [Crossref] [PubMed]
- Yamamoto H, Watanabe K, Nagata M, et al. Surgical treatment for pancreatic metastasis from soft-tissue sarcoma: report of two cases. Am J Clin Oncol 2001;24:198-200. [Crossref] [PubMed]
- Luc G, Collet D, Reich S, et al. Primary monophasic synovial sarcoma of the pancreas. J Visc Surg 2013;150:159-61. [Crossref] [PubMed]
- Subramaniam MM, Noguera R, Piqueras M, et al. p16INK4A (CDKN2A) gene deletion is a frequent genetic event in synovial sarcomas. Am J Clin Pathol 2006;126:866-74. [Crossref] [PubMed]
- Sabah M, Cummins R, Leader M, et al. Loss of p16INK4A expression is associated with allelic imbalance/loss of heterozygosity of chromosome 9p21 in microdissected synovial sarcomas. Virchows Arch 2005;447:842-8. [Crossref] [PubMed]



