
The decreased expression of hsa_circ_0043278 and its relationship with clinicopathological features of breast cancer
Introduction
Breast cancer is one of the most common cancers worldwide and is a significant cause of death in women (1,2). Although comprehensive treatment strategies and the diagnosis of breast cancer have improved significantly, local recurrence and distant metastasis still occur frequently. The absence of biomarkers for early detection is one reason for the poor prognosis. Therefore, the exploration of new biomarkers and therapeutic targets for breast cancer treatment is urgently needed.
Compared with linear RNAs, circular RNAs (circRNAs) are stable and conserved RNA molecules (3,4). CircRNAs act as sponges of micro RNAs (miRNAs), thus interfering with the expression of their target mRNAs, and are associated with the development of multiple human diseases (3,5-9). Accumulating evidence shows circRNAs play pivotal roles in cancer biology. Li et al. (10) suggested circDDX17 functioned as a tumor suppressor in colorectal cancer. Circ_0030235 might play a crucial role in human pancreatic ductal adenocarcinoma by sponging miR-1253 and miR-1294 (11). In human breast cancer, hsa_circ_0001982 affected breast cancer cell proliferation, invasion, and induced apoptosis by targeting miR-143 (12). Gao et al. (13) suggested that circ_0006528 promoted breast cancer growth, invasion, and migration through the circ_0006528/miR-7-5p/Raf1/MEK/ERK network in breast cancer. However, few reports have been published on the role of circRNAs in human breast cancer.
Here, the results of our study showed that hsa_circ_0043278 was downregulated in breast cancer tissues using a microarray. The expression of hsa_circ_0043278 was verified using quantitative real-time polymerase chain reaction (qRT-PCR). The relationship of hsa_circ_0043278 with the clinicopathological characteristics in human breast cancer was confirmed. This study supplies evidence that hsa_circ_0043278 may play a key role and might be a potential marker for human breast cancer. We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/gs-20-825).
Methods
Samples and clinicopathological information
The clinicopathological information of 50 breast cancer patients and 38 benign breast tumor patients was collected from the First Affiliated Hospital of China Medical University (from January 2005 to December 2012). The clinicopathological information, including age, tumor size, lymph node metastasis, histological type, histological grade, estrogen receptor (ER) status, progesterone receptor (PR) status, human epidermal growth factor receptor 2 (HER-2, ErbB-2) status, and Ki67 expression was collected. The expression of ER, PR, HER-2, and Ki67 were following the pathological results. Fresh breast cancer tissues and paired noncancerous tissues were retrieved from 50 patients who underwent surgical treatment. The paired noncancerous tissues were collected over 5 cm from the edge of the cancer tissues. Benign breast tumor tissues were collected from 38 patients, including 28 cases of fibroadenoma, 7 cases of intraductal papilloma, and 3 cases of hyperplastic nodules. The samples were stored at −80 °C.
All malignant clinical data samples were primary female breast cancer (from 27 to 81 years old), and none of the patients had received any therapeutic interventions, including chemotherapy or radiotherapy. The study was approved by the Ethics Committee of The First Affiliated Hospital of China Medical University (No. AF-0G-03-1.0-02). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent was retrieved from all participants in the study.
circRNA microarray
The Arraystar Human circRNA Microarray (KangChen Biotech, Shanghai, China), which covered 2465 human circRNAs, was used to identify the differential expression of circRNAs among three pairs of breast cancer tissues and paired noncancerous tissues. The total number of RNA samples was quantified by NanoDrop ND-1000 (Thermo, USA). The samples were subjected to microarray hybridization according to Arraystar’s standard protocols. Differentially expressed circRNAs were found, and the images and data were collected and analyzed by software.
QRT-PCR
We verified hsa_circ_0043278 expression by qRT-PCR in 50 breast cancer tissues and paired noncancerous tissues, and 38 benign breast tumor tissues as control. Total RNA was extracted by TRIZOL reagent (Takara Bio, Japan) following the manufacturer’s instructions. Each of the cDNAs was reverse transcribed from 2 µg of total RNA using a PrimeScriptTM RT reagent kit with gDNA eraser (Perfect Real Time) reverse transcriptase (Takara Bio). SYBR performed the reaction method-based qRT-PCR using SYBR®Premix Ex TaqTMII (Takara Bio). A Light Cycler 480 II sequence detection system (Roche Applied Science, Indianapolis, USA). The hsa_circ_0043278 RNA expression levels were normalized to those of GAPDH. The hsa_circ_0043278 and GAPDH primers are shown in Table 1. The relative expression of hsa_circ_0043278 is analyzed using 2−ΔCT value. The experiment was repeated thrice.

Full table
Bioinformatics prediction
The target miRNAs of hsa_circ_0043278 are predicted by miRNA target prediction software, including TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/). The top 5 of the target miRNAs were selected from the result of the prediction.
Statistical analysis
Statistical data was analyzed using GraphPad Prism 7.0 software (GraphPad Software, LaJolla, CA, USA). One-way ANOVA was used to verify the circRNA expression in breast cancer tissues, paired noncancerous tissues, and benign breast tumors. Kruskal-Wallis test and Mann-Whitney test were performed for the association between circRNA expression and clinicopathological data. The results for qRT-PCR were presented as mean ± SEM, and the other data were presented as mean ± SD. The receiver operating characteristic (ROC) curve was constructed by Statistical Product and Service Solutions (SPSS) 19.0 software (SPSS, Chicago, IL, USA). A P value <0.05 was considered statistically significant.
Results
Differentially expressed circRNAs screened by microarray
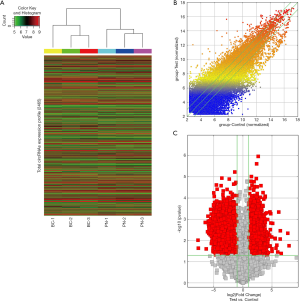
A total of 2,465 human circRNAs in three breast cancer tissues and paired noncancerous tissues were screened by microarray (Figure 1A). The expression pattern of all circRNAs in breast cancer tissues and paired noncancerous tissues was shown by hierarchical clustering (Figure 1A). Furthermore, 520 differentially expressed circRNAs, including 292 upregulated and 228 downregulated circRNAs in breast cancer tissues, were detected according to the filter criteria (fold-change ≥2 and P<0.05). The distribution of the differential expressed circRNAs between breast cancer tissues and paired noncancerous tissues was evaluated by a scatter plot (Figure 1B) and a volcano plot (Figure 1C).
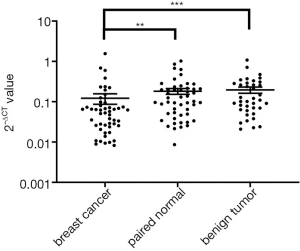
Validation of hsa_circ_0043278 in breast cancer tissues by qRT-PCR
According to the microarray results, a novel circRNA hsa_circ_0043278 downregulated 43-fold in breast cancer was chosen for further validation by qRT-PCR. It is located at chromosome 17, and its gene symbol is TADA2A. The result showed that the expression of hsa_circ_0043278 was downregulated in 68% (34/50) of breast cancer tissues compared to paired noncancerous tissues. As shown in Figure 2, the expression level of hsa_circ_0043278 is significantly downregulated in breast cancer tissues compared with paired noncancerous tissues (P<0.01). This result was consistent with the microarray and suggested hsa_circ_0043278 might play a role in the occurrence of breast cancer.
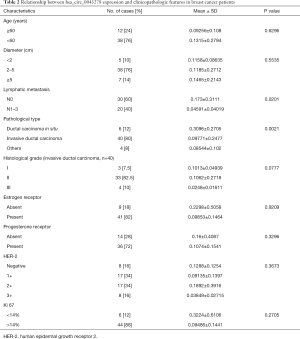
The relationship between hsa_circ_0043278 levels and clinicopathological characteristics
Furthermore, the relationship between hsa_circ_0043278 expression levels and the clinicopathological characteristics was examined. As shown in Table 2, the results demonstrated that the expression of hsa_circ_0043278 was associated with lymph node metastasis and histological type of the patient. Patients with lymph node metastasis have tumors with significantly downregulated expression of hsa_circ_0043278 (P=0.0201). Also, the expression levels of hsa_circ_0043278 are significantly downregulated in invasive ductal carcinoma compared with ductal carcinoma in situ (P=0.0021). However, the expression levels of hsa_circ_0043278 have no relationship with other clinicopathological characteristics.
Full table
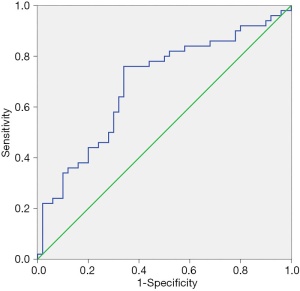
ROC curve of hsa_circ_0043278
A ROC curve was constructed to evaluate the diagnostic value of hsa_circ_0043278 in breast cancer. The area under the ROC curve was 0.690 [95% confidence interval (CI) =0.585–0.795; P=0.001; Figure 3]. It suggested that hsa_circ_0043278 might have a potential impact on the occurrence of breast cancer.
Prediction of hsa_circ_0043278 target genes
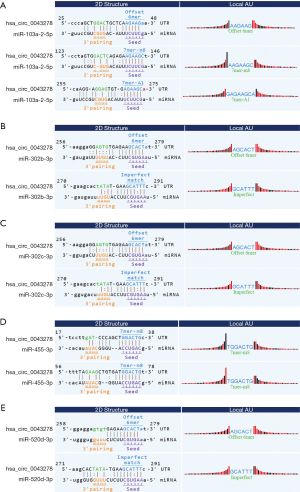
To find the target miRNAs, we used bioinformatics tools to predict the target miRNAs of hsa_circ_0043278. As shown in Figure 4, we have selected five miRNAs likely to match hsa_circ_0043278: miR-103a-2-5p, miR-302b-3p, miR-302c-3p, miR-455-3p, and miR-520d-3p. The possible target sites were then predicted, and the binding sites were represented by vertical solid lines (Figure 4). The result suggests hsa_circ_0043278 possibly functions through these target miRNAs.
Discussion
circRNAs are a particular type of conserved RNA and are a popular topic in the field of RNA research. However, at present, our understanding of circRNA is still limited (14,15). Few circRNAs have been reported in breast cancer. In our paper, downregulated circRNA hsa_circ_0043278 was found using a microarray and verified using qRT-PCR in breast cancer tissues. Further research revealed that the expression of hsa_circ_0043278 was correlated with histological type and lymph node metastasis of breast cancer. The results suggested that hsa_circ_0043278 played a role in breast cancer and had a potential research value.
Hsa_circ_0043278 was downregulated 43-fold in breast cancer tissues, and its biological function has not yet been reported. The results of microarray and qRT-PCR suggested hsa_circ_0043278 might be related to the progression of breast cancer. However, due to the low expression level of circRNAs in the human body, it is difficult to avoid the potential for false negatives in evaluating circRNA expression. Further experiments are needed to verify these findings.
Consequently, we found that the expression of hsa_circ_0043278 was much lower in patients with greater lymph node involvement and poorly differentiated breast cancer. Despite the limitation in the sample size in this study, our results showed that hsa_circ_0043278 expression might be correlated with lymph node invasion and the degree of malignancy of breast cancer. Our results showed that low expression of hsa_circ_0043278 might be correlated with a poorer prognosis in breast cancer. These findings will need further verification.
Tumor biomarkers and their combined detection are beneficial for the screening of tumors, but few reports of tumor biomarkers in breast cancer have been published. In our study, the ROC curve was constructed, and the result showed that the AUC was 0.690, which indicates that hsa_circ_0043278 might have potential diagnostic value for breast cancer. However, we need to expand the sample size for further verification.
MiRNAs can bind and inhibit 3’UTR activity of target mRNAs from repressing these coding genes (16-20). It is generally recognized that circRNAs act as potent miRNA sponges (5) and serves as competing endogenous RNAs (21,22). Therefore, we predicted five target miRNAs of hsa_circ_0043278 using bioinformatics tools. Some miRNAs were found to be associated with breast cancer. Wang (23) revealed miR-455 was downregulated in breast cancer tissues and cells, and that it inhibited cell proliferation by targeting CDK14. This result showed that miR-455 could be a target for breast cancer therapy. MiR-455-3p was reported to play an essential role in melanoma and could thus be a potential target for treatment (24). Preliminary research has indicated that miR-302b and miR-302c were associated with the occurrence and development of breast cancer (25). MiR-302b could enhance the sensitivity of breast cancer cells to cisplatin (26), while miR-302a/b/c/d could also sensitize breast cancer cells to Adriamycin (27). Another analysis predicted that miR-520d and miR-302c corresponded with HER2/neu (28). Therefore, we hypothesize that hsa_circ_0043278 might have a role in breast cancer and functions via the regulation of these putative target miRNAs. Nonetheless, further research is needed to verify the interaction between hsa_circ_0043278 and these miRNAs. The expression of these five miRNAs in breast cancer also requires further confirmation.
In summary, circRNAs were found to be highly prevalent spliced transcripts from hundreds of genes (21), and a subset of them are related to multiple malignancies. We detected differentially expressed circRNAs in breast tissue samples by microarray and determined hsa_circ_0043278 was downregulated in breast cancer. The results suggest that hsa_circ_0043278 may play a role in breast cancer and could be a promising marker for breast cancer diagnosis. Nonetheless, how hsa_circ_0043278 affects the biological behavior of breast cancer needs to be further investigated. It will also be essential to determine how hsa_circ_0043278 can be used for future breast cancer screening and treatment.
Acknowledgments
We thank The First Affiliated Hospital of China Medical University for technical support.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/gs-20-825
Data Sharing Statement: Available at http://dx.doi.org/10.21037/gs-20-825
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/gs-20-825). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics Committee of The First Affiliated Hospital of China Medical University (No. AF-0G-03-1.0-02). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent was retrieved from all participants in the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Parrella P. Epigenetic Signatures in Breast Cancer: Clinical Perspective. Breast Care (Basel) 2010;5:66-73. [Crossref] [PubMed]
- Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin 2012;62:220-41. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Moirangthem A, Patel T. Circular RNA: non-coding RNA with emerging roles in stem cell differentiation. Transl Cancer Res 2018;7:S50-2. [Crossref]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Meng X, Li X, Zhang P, et al. Circular RNA: an emerging key player in RNA world. Brief Bioinform 2017;18:547-57. [PubMed]
- Shao Y, Chen Y. Roles of Circular RNAs in Neurologic Disease. Front Mol Neurosci 2016;9:25. [Crossref] [PubMed]
- Fittipaldi S, Vasuri F. circRNAs and miRNAs in the diagnosis of hepatocellular carcinoma: the long race to new targets. Transl Cancer Res 2018;7:S726-9. [Crossref]
- Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer 2019;18:20. [Crossref] [PubMed]
- Li XN, Wang ZJ, Ye CX, et al. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res 2018;37:325. [Crossref] [PubMed]
- Xu Y, Yao Y, Gao P, et al. Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294. Biochem Biophys Res Commun 2019;509:138-42. [Crossref] [PubMed]
- Tang YY, Zhao P, Zou TN, et al. Circular RNA hsa_circ_0001982 Promotes Breast Cancer Cell Carcinogenesis Through Decreasing miR-143. DNA Cell Biol 2017;36:901-8. [Crossref] [PubMed]
- Gao D, Qi X, Zhang X, et al. hsa_circRNA_0006528 as a competing endogenous RNA promotes human breast cancer progression by sponging miR-7-5p and activating the MAPK/ERK signaling pathway. Mol Carcinog 2019;58:554-64. [Crossref] [PubMed]
- Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014;15:409. [Crossref] [PubMed]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. [Crossref] [PubMed]
- Ambros V. The functions of animal microRNAs. Nature 2004;431:350-5. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell 2006;11:441-50. [Crossref] [PubMed]
- Selbach M, Schwanhausser B, Thierfelder N, et al. Widespread changes in protein synthesis induced by microRNAs. Nature 2008;455:58-63. [Crossref] [PubMed]
- Shukla GC, Singh J, Barik S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol Cell Pharmacol 2011;3:83-92. [PubMed]
- Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733. [Crossref] [PubMed]
- Xu H, Guo S, Li W, et al. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep 2015;5:12453. [Crossref] [PubMed]
- Wang B, Zou A, Ma L, et al. miR-455 inhibits breast cancer cell proliferation through targeting CDK14. Eur J Pharmacol 2017;807:138-43. [Crossref] [PubMed]
- Chai L, Kang XJ, Sun ZZ, et al. MiR-497-5p, miR-195-5p and miR-455-3p function as tumor suppressors by targeting hTERT in melanoma A375 cells. Cancer Manag Res 2018;10:989-1003. [Crossref] [PubMed]
- Chen D, Yang H. Integrated analysis of differentially expressed genes in breast cancer pathogenesis. Oncol Lett 2015;9:2560-6. [Crossref] [PubMed]
- Cataldo A, Cheung DG, Balsari A, et al. miR-302b enhances breast cancer cell sensitivity to cisplatin by regulating E2F1 and the cellular DNA damage response. Oncotarget 2016;7:786-97. [Crossref] [PubMed]
- Zhao L, Wang Y, Jiang L, et al. MiR-302a/b/c/d cooperatively sensitizes breast cancer cells to adriamycin via suppressing P-glycoprotein(P-gp) by targeting MAP/ERK kinase kinase 1 (MEKK1). J Exp Clin Cancer Res 2016;35:25. [Crossref] [PubMed]
- Lowery AJ, Miller N, Devaney A, et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res 2009;11:R27. [Crossref] [PubMed]
(English Language Editors: Fitzgerald and J. Chapnick)


