
Caveolin-1 regulates the expression of miR-183 and inhibits the invasion and migration of invasive pituitary adenomas by affecting early growth response 1 (EGR1)/Krueppel-like factor 5 (KLF5) interaction
Introduction
Pituitary adenoma (PA) is the most common benign endocrine tumor of the central nervous system, accounting for about 15% of all primary intracranial tumors (1). Current clinical studies have shown that approximately 45–55% of pituitary adenomas will invade the cavernous sinus, sphenoid sinus, internal carotid artery, and other important brain tissues. The tumor tissue cannot be completely removed by surgery, and the recurrence rate is extremely high (2); this type of pituitary adenoma is known as invasive pituitary adenoma (IPA). A clear understanding of the pathogenesis of IPA is critical for better control of its occurrence and development. Caveolin-1 (CAV-1) is an integral membrane protein. It is also the main component of microvesicles, and plays a role in the transportation and signal transduction of microvesicles (3). Studies have shown that the expression of CAV-1 is up-regulated in a variety of tumors, especially in aggressive tumors (3). An increasing number of studies have shown that the up-regulation of CAV-1 expression enhances the invasion and migration ability of tumor cells, however its exact function in IPA is still unknown. Lee et al. has shown that CAV-1 can selectively regulate the expression of micro ribonucleic acids (miRNAs) through microbubbles (4). MiRNA is an endogenous non-coding single-stranded RNA that is widely distributed in eukaryotic cells, which can regulate the translation and expression of target genes by binding to the 3'-untranslated region (UTR) region of target genes. Abnormal expression of miRNA is often related to the proliferation, invasion, migration, and cell cycle of cancer cells. Duan et al. reported that the expression of miRNA (miR)-137 in IPA tissue is severely lacking compared with normal pituitary gland tissue. Up-regulating the target of miR-137 significantly inhibits the proliferation and migration of IPA cells (5). Meanwhile, other studies have also pointed out that a variety of miRNAs are differentially expressed in IPA tissues or cells, including miR-16 (6), miR-106b (7), miR-212 (8), miR-133 (9), and miR-410 (10). It has been found that the expression of miR-183 is imbalanced in a variety of nervous system tumor tissues or cells, which affects the invasion and migration ability of cancer cells (11-13). However, the expression of miR-183 in IPA tissues and the regulatory effect of miR-183 on the biological functions of IPA cells have not yet been elucidated in the existing literature.
In this study, we will examine the expression levels of CAV-1 and miR-183 in IPA tissues and cells, explore the possible regulatory mechanisms between them, and study their impacts on the invasion and migration of IPA cells at the cellular level to identify new methods for the treatment of IPA.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/gs-20-823).
Methods
Cells and materials
Pathological tissue
One hundred and forty-three cases of pituitary tumor tissue samples obtained during pituitary adenoma resection in our hospital from March 2017 to December 2019 were collected. The samples were divided into invasive pituitary tumor (IPA) tissue and non-invasive pituitary adenoma (non-IPA) tissue according to the postoperative pathological diagnosis. This study was approved by the ethics committee of our hospital, and written informed consent was obtained from the patients and their families. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Cells
Rat pituitary adenoma GH3 cell line and rat pituitary RPC cell line were purchased from Shanghai Tongwei Biotechnology Co. Ltd. (Shanghai, China), and DH5α Escherichia coli (E. coli) strain was purchased from Beijing Biolab Technology Co. Ltd. (Beijing, China).
Experimental materials
The phosphate-buffered saline (PBS) buffer, pancreatin, penicillin, streptomycin, fetal bovine serum, skimmed milk powder, L-glutamine, and Dulbecco’s Modified Eagle Medium (DMEM) medium were purchased from MP Biomedicals (California, USA). The Lipofectamine 3000 transfection kit and Trizol total RNA extraction kit were purchased from Invitrogen (California, USA). The radioimmunoprecipitation assay (RIPA) lysate, protein assay [bicinchoninic acid (BCA)] kit, and enhanced chemiluminescence (ECL) detection kit were purchased from Shanghai Jizhi Biochemical Technology Co. Ltd. (Shanghai, China). The nuclear/cytoplasmic protein separation kit was purchased from PromoCell (Heidelberg, Germany). The CAV-1 polyclonal antibody, early growth response 1 (EGR1) monoclonal antibody, Krueppel-like factor 5 (KLF5) polyclonal antibody, and ezrin (EZR) polyclonal antibody were purchased from Wuhan Amictech Technology Co. Ltd. (Wuhan, China). The SYBR GREEN kit was purchased from China Sino Biological. Inc (Beijing, China). The dual-luciferase reporter gene experiment kit was purchased from Abcam, (Cambridge, UK). The Transwell chamber and crystal violet staining solution were purchased from Corning (New York, USA).
Cell culture
The IPA tumor tissue and non-IPA tissue were washed three times with PBS and cut into small pieces (1 mm3). Subsequently, the tissue was digested with 0.25% pancreatin at 37 °C, and cultured in DMEM containing 10% fetal calf serum. Thirty minutes later, DMEM medium containing 10% fetal bovine serum was added to terminate the digestion. After centrifugation at 1,500 rpm for 5 min, the cells were collected and resuspended in DMEM medium containing 20% fetal bovine serum. The medium was changed every 2 days.
The rat pituitary adenoma GH3 cell line was inoculated in DMEM medium containing 10% fetal bovine serum, 100 U/mL penicillin, 100 U/mL streptomycin, and 0.03% L-glutamine, and cultivated in a constant temperature incubator at 37 °C with 5% carbon dioxide (CO2) for subsequent experiments.
Construction and transfection of lentivirus
The short hairpin RNA (shRNA) targeting CAV-1 and EZR, and its blank control RNA (sh-Ctrl), were cloned into pGCSIL-GFP plasmid. After being transferred to DH5α E. coli, the recombinant plasmid was extracted and transfected into GH3 cells using the Lipofectamine 3000 kit. The experiment was performed 48 h after transfection. The GH3 cells in the logarithmic growth phase were used, and the cell concentration was adjusted to 1×105 cells/mL after trypsinization treatment. Next, the cells were inoculated in a 6-well plate with a volume of 2 mL per well, which was subsequently cultivated in a constant temperature incubator at 37 °C with 5% CO2 for 24 h.
Extraction of EGR1 and KLF5 proteins in GH3 cytoplasm and nucleus
The RIPA lysate was used to extract the total protein in tissue and GH3 cells. After centrifugation at 3,000 rpm for 30 min, the supernatant was collected. The cytoplasmic and nuclear proteins were then separated according to the instructions of the nuclear/cytoplasmic protein separation kit. Finally, the protein concentration was determined by the BCA method.
Western blot experiment to detect the expression of CAV-1, EGR1, KLF5, and EZR in tissues and cells
RIPA lysate was used to extract the total protein in tissue and GH3 cells, and the BCA method was used to determine total protein concentration. The protein was separated by adding polyacrylamide gel, and was then transferred to the polyvinylidene fluoride (PVDF) membrane. After blocking with 5% skimmed milk powder for 1 h, diluted primary antibody (1:1,000) was added and incubated overnight at 4 °C. The primary antibody was removed the next day, and rinsed with Tris Buffered saline Tween (TBST) three times. Following this, the diluted secondary antibody (1:5,000) was added and incubated at room temperature for 1 h. After rinsing with TBST three times, ECL luminescent solution was added for development. Finally, after taking pictures, image J software National Institutes of Health Company (Maryland, USA) was used to analyze the gray value of each band.
Quantitative real-time polymerase chain reaction (qRT-PCR) detects the expression of miR-183 in IPA tissues and GH3 cells
After collecting the cells, the Trizol kit was used to extract the total RNA in the cells, and the RNA concentration was detected. The extracted RNA was reverse transcribed into complementary deoxyribonucleic acid (cDNA) using a reverse transcription kit, and U6 was used as an internal reference to design the primer sequence of miR-183 (F: 5'-GCGAGCACAGAATTA ATACGACTCACTATAGG-3'; R: 5'-ATGGCACTGGTAGAATTCACT-3'). The PCR reaction system was constructed with a final volume of 20 µL [2 µL reverse transcription product, 10 µL SYBR Green Mix, 0.5 µL upstream and downstream primers, and 7 µL distilled water (dH2O) each] by following the SYBR GREEN kit instructions to detect the expression level of miR-183. The parameters of the PCR reaction system were as follows: pre-denaturation at 95 °C for 5 min, denaturation at 94 °C for 30 s, and annealing at 60 °C for 30 s. The reaction was carried out for a total of 45 cycles. The results were calculated using the 2−ΔΔCt method.
The detection of luciferase activity in GH3 cells by double luciferase reporter gene experiment
After amplifying EGR1 and KLF5, they were cloned into pcDNA3.1 plasmid, and the miR-183 promoter was amplified using specific primers and inserted into the pGL-3 luciferase reporter vector to construct pGL3-rno-miR-183-luc. GH3 cells were inoculated in a 9-well plate at a density of 2×104 cells/well. When the cells were fused to approximately 80%, the Lipofectamine 3000 kit was used to co-transfect pc-EGR1 or pc-KLF5 with pGL3-rno-miR-183-luc into GH3 cells, and a dual luciferase assay system was used to detect intracellular luciferase activity 24 h after transfection.
The wild-type EZR (WT-EZR)-3'-UTR and mutant EZR (MUT-EZR)-3’-UTR regions were cloned to the downstream of the pGLO-basic firefly luciferase gene, and the plasmid was transfected with miR-negative control (NC) or miR-183 mimics using the Lipofectamine 3000 kit into GH3 cells, and a dual luciferase assay system was used to detect intracellular luciferase activity 24 h after transfection.
Co-immunoprecipitation test to detect the interaction between EGR1 and KLF5
GH3 cells in the logarithmic growth phase were collected, and an appropriate amount of RIPA lysis buffer was added and lysed at 4 °C for 30 min. Next, the supernatant was collected after centrifuging at 3,000 rpm for 30 min. G-Sepharose protein was used for pretreatment. The collected supernatant with the indicator antibody were incubated overnight at 4 °C. Four multiplication Sodium dodecyl sulfate-Polyacrylamide gel electrophoresis (SDS-PAGE) buffer was added after the immunoprecipitation reaction and boiled in water for 10 min. Finally, western blotting was performed to obtain the binding protein for subsequent analysis.
Detection of GH3 cell invasion ability with Transwell test
GH3 cells were inoculated on the upper layer of the Transwell cell covered with matrigel, and serum-free DMEM medium was added. DMEM medium containing 5% fetal bovine serum was added to the lower layer of the Transwell chamber. After incubating for 48 h in a constant temperature incubator, the upper layer of cells that did not invade was washed with PBS and stained with crystal violet. Finally, pictures were taken, and cells were counted to evaluate their invasion ability.
Cell scratch test to detect the migration ability of GH3 cells
GH3 cells were inoculated into a 6-well plate at a density of 5×105 cells/well. After incubating overnight in a constant temperature incubator, the cell layer was scraped with a sterile pipette tip, and the remaining cell debris was rinsed with fresh medium. The complete medium was then added to the 6-well plate and incubated for 48 h. Following incubation, a picture was taken under the microscope and the distance between the scratches was recorded.
Statistical analysis
All experiments in this study were repeated three times, and the average value ± standard deviation (SD) was used. SPSS 22.0 from IBM (New York, USA) was used for statistical analysis. The t-test was used to evaluate the difference between the two groups, and one-way analysis of variance (ANOVA) was used to evaluate the differences in data between multiple groups. P<0.05 indicated that the difference was statistically significant, and all graphs were drawn using GraphPad 8.2.1from GraphPad Software company (California, USA).
Results
Down-regulation of CAV-1 inhibits GH3 cell invasion and migration
First, the expression of CAV-1 in IPA tissue and non-IPA tissue was detected by Western blot. The results showed that the expression of CAV-1 in IPA tissue was significantly up-regulated (P<0.05, Figure 1A). At the same time, the expression of CAV-1 in GH3 and RPC cells was also detected, and the results showed that CAV-1 was highly expressed in GH3 cells (P<0.05, Figure 1B). The expression of CAV-1 in GH3 cells was down-regulated by the transfection of lentivirus (P<0.05), and it was found that down-regulating the expression of CAV-1 can significantly inhibit the invasion and migration ability of GH3 cells (P<0.05, Figure 1C,D,E).
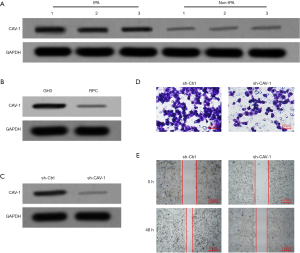
Down-regulation of CAV-1 inhibits EGR1/KLF5 interaction and up-regulates the expression of miR-183
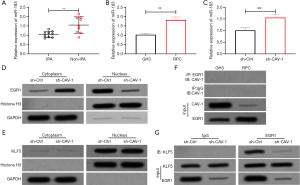
The expression level of miR-183 in IPA tissues and cells was detected by qRT-PCR. The results showed that the expression of miR-183 in IPA tissues and GH3 cells was significantly absent (P<0.01, Figure 2A,B). After inhibiting the expression of CAV-1 in GH3 cells, the expression level of miR-183 was notably restored (P<0.01, Figure 2C). Western blot results showed that after down-regulating the expression of CAV-1, the expression of EGR1 in the GH3 nucleus was markedly reduced (P<0.05), however, down-regulating the expression of CAV-1 did not affect the expression of KLF5 in the cytoplasm and nucleus (P>0.05, Figure 2D,E). The interaction between CAV-1 and EGR1 was further clarified by immunoprecipitation. The results showed that CAV-1 did not bind to EGR1 in GH3 and RPC cells, but down-regulating the expression of CAV-1 would inhibit the combination of EGR-1 with KLF5 (P<0.05, Figure 2F,G).
EGR-1 inhibits the transcriptional activation of miR-183 by interacting with KLF5
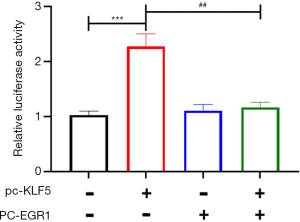
The luciferase report experiment was conducted to verify whether the interaction between EGR1 and KLF5 affects the transcriptional activation of miR-183. The results showed that the luciferase activity of miR-183 was significantly increased after KLF5 was transfected alone (P<0.001). However, transfection of EGR1 alone did not affect the luciferase activity of miR-183. Moreover, co-transfection of EGR1 and KLF5 into GH3 cells notably inhibited the increase in miR-183 luciferase activity mediated by KLF5 (P<0.05, Figure 3).
EZR is a downstream target gene of miR-183
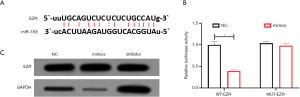
The prediction results from the bioinformatics website StarBase showed that EZR and miR-183 have partial target binding sequences in 3'-UTR (as shown in Figure 4A). At the same time, the results of the dual-luciferase reporter gene experiment showed that miR-29a can significantly inhibit the luciferase activity of WT-EZR (P<0.05), but has no obvious inhibitory effect on the MUT-EZR luciferase activity (as shown in Figure 4B). Western blot experiments showed that inhibiting the expression of miR-183 would up-regulate the expression of EZR in GH3 cells (P<0.05), and overexpression of miR-183 would inhibit the expression of EZR in the cells (P<0.05, Figure 4C).
Effects of miR-183 and EZR on the invasion and migration of GH3 cells
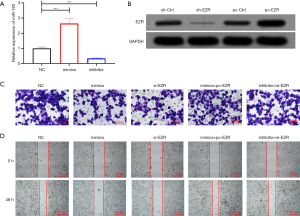
QRT-PCR was used to detect the expression level of miR-183 in GH3 cell. The expression level of miR-183 in the cell was markedly increased after the transfection of miR-183 mimics (P<0.001), and was notably suppressed after the transfection of miR-183 inhibitor (P<0.001, Figure 5A). Western blot experiment results showed that the expression of the EZR protein in cells was significantly reduced after sh-EZR transfection (P<0.05), but was substantially increased after transfection of pc-EZR (P<0.05, Figure 5B). The invasion and migration ability of GH3 cells was detected by cell invasion experiments and the cell scratch experiment. The results showed that overexpression of miR-183 or inhibition of EZR expression significantly inhibited the invasion and migration ability of GH3 cells (P<0.05), however, the simultaneous overexpression or inhibition of miR-183 and EZR expression exhibited no obvious effect on the invasion and migration ability of GH3 cells (Figure 5C,D).
Discussion
The CAV-1 gene is related to cell metastasis and has been confirmed to play a role in numerous tumors. Torrejón et al. confirmed that the expression of CAV-1 in patients with colorectal cancer is usually down-regulated, and exerts a tumor suppressor effect in the early stages of the disease (14). Similarly, CAV-1 also exerts a tumor suppressor effect in breast and ovarian cancers (15,16). However, some studies report that CAV-1 accelerates tumor progression in prostate, bladder, and pancreatic cancers (17-19). Liu et al. showed that the down-regulation of CAV-1 in the early stage of lung cancer promotes the proliferation of cancer cells, but the up-regulation of CAV-1 in the late stage gives lung cancer cells a greater ability to invade (20). In the present study, it was found that the expression of CAV-1 in IPA tissue was significantly up-regulated compared with non-IPA tissue. Additionally, the up-regulation of CAV-1 expression was also found in GH3 cells, which indicates that the up-regulation of CAV-1 expression may enhance the invasion and migration of pituitary adenoma cells. By knocking down CAV-1 to inhibit its expression in GH3 cells, it was found that inhibiting CAV-1 expression can effectively reduce the ability of GH3 cells to invade and migrate, confirming the above findings.
CAV-1 is the main component of microvesicles, which can selectively regulate the expression level of miRNAs through vesicles. In previous experiments, it was found that inhibiting the expression of CAV-1 could significantly up-regulate the expression level of miR-183 in GH3 cells. MiR-183 is a member of the miR-183-96-182 family located at 7q31-34 of human chromosome 5, and is highly conserved in different organisms (21). Furthermore, it is abnormally expressed in many types of cancers and participates in the occurrence and development of tumors. Studies have shown that the expression of miR-183 is up-regulated in 18 cancers, including prostate, ovarian, and breast cancers, and exhibits carcinogenic effects. However, the expression of miR-183 is down-regulated in six cancers, including osteosarcoma and melanoma, and exerts a tumor suppressor effect (21). Cao et al. reported that eight regulatory factors can control the expression of miR-183 by combining with the promoter region on it (21). In the present study, it was found that the expression of miR-183 in IPA tissues and GH3 cells was significantly reduced, but knocking down CAV-1 up-regulated the expression of miR-183 in GH3 cells; that is, the lack of miR-183 may lead to the enhancement of GH3 cell invasion and migration. Studies have shown that CAV-1 can affect the transcriptional activation of miRNAs by affecting nuclear ectopic transcription factors, thereby regulating the expression of miRNAs, including EGR1 and KLF5. The results of the present study showed that the expression of EGR1 in the nucleus of GH3 cells decreased after knocking down CAV-1, and this may be caused by the lack of Cav-1 expression inhibiting its binding to the promoter on the common site of EGR1 protein Sp1 (22). However the expression of KLF5 had no significant changes after knocking down CAV-1. It was also confirmed by immunoprecipitation experiments that knockdown of CAV-1 substantially inhibited the interaction between EGR1 and KLF5; that is, knocking down CAV-1 can hinder the formation of the EGR1/KLF5 complex by reducing the ectopic core of EGR1, thereby producing a high concentration of free KLF5. Qin et al. showed that KLF5 can actively regulate the expression of miRNAs (such as miR-183) directly through combination with the promoter (22). The luciferase report experiment confirmed that up-regulating KLF5 can considerably enhance the luciferase activity in GH3 cells, but up-regulating EGR1 exhibits no significant effect on luciferase activity. Moreover, simultaneously upregulating EGR1 and KLF5 has no obvious impact on the intracellular luciferase activity. The above results indicate that CAV-1 can regulate the expression of miR-183 by affecting the interaction between EGR1 and KLF5.
Numerous studies have shown that the lack of expression of miR-183 plays an important role in the ability of tumor cells to invade and migrate. miR-183 can regulate approximately 45 tumor-related genes (such as EZR), which are closely related to the adhesion, invasion, and migration capabilities of tumor cells (21). EZR, also known as Villin2 or Cytovillin2, has been confirmed to be involved in the metastasis and proliferation of a variety of tumors (23). In a recent study, EZR was listed as a new type of biomarker for IPA, and cell experiments have found that inhibiting the expression of EZR can reduce the invasion and migration ability of GH3 cells (24). In the present study, it was found that EZR and miR-183 have partial targeting binding sequences in 3'-UTR, and miR-183 can negatively regulate the expression of EZR in GH3 cells. The dual-luciferase reporter gene experiment also confirmed the targeting relationship between EZR and miR-183. At the same time, overexpression of miR-183 or inhibition of EZR expression can significantly inhibit the invasion and migration of GH3 cells. However, the simultaneous overexpression or inhibition of miR-183 and EZR expression has no obvious effect on the invasion and migration of GH3 cells.
In conclusion, the high expression of CAV-1 in IPA tissues and GH3 cells may reduce the expression of miR-183 by promoting the interaction of EGR1 and KLF5, thereby strengthening the invasion and migration capabilities of pituitary adenoma tissues and GH3 cells. Although CAV-1 affects the nuclear ectopic of EGR1, no direct correlation between CAV-1 and EGR1 was identified in this study. The mechanism of interaction between CAV-1 and EGR1 requires further investigation in future studies.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/gs-20-823
Data Sharing Statement: Available at http://dx.doi.org/10.21037/gs-20-823
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/gs-20-823). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the ethics committee of our hospital (No. ZXYY-KY-0148), and written informed consent was obtained from the patients and their families. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhu H, Miao Y, Shen Y, et al. The clinical characteristics and molecular mechanism of pituitary adenoma associated with meningioma. J Transl Med 2019;17:354. [Crossref] [PubMed]
- Wang X, Zhang D, Ma S, et al. Predicting the likelihood of early recurrence based on mRNA sequencing of pituitary adenomas. Gland Surg 2019;8:648-56. [Crossref] [PubMed]
- Ketteler J, Klein D. Caveolin-1, cancer and therapy resistance. Int J Cancer 2018;143:2092-104. [Crossref] [PubMed]
- Lee H, Li C, Zhang Y, et al. Caveolin-1 selectively regulates microRNA sorting into microvesicles after noxious stimuli. J Exp Med 2019;216:2202-20. [Crossref] [PubMed]
- Duan J, Lu G, Li Y, et al. miR-137 functions as a tumor suppressor gene in pituitary adenoma by targeting AKT2. Int J Clin Exp Pathol 2019;12:1557-64. [PubMed]
- Wang DW, Wang YQ, Shu HS. MiR-16 inhibits pituitary adenoma cell proliferation via the suppression of ERK/MAPK signal pathway. Eur Rev Med Pharmacol Sci 2018;22:1241-48. [PubMed]
- Zheng Z, Zhang Y, Zhang Z, et al. Effect of miR-106b on Invasiveness of Pituitary Adenoma via PTEN-PI3K/AKT. Med Sci Monit 2017;23:1277-85. [Crossref] [PubMed]
- Wang HY, Gao HQ. Reduction of miR-212 contributes to pituitary adenoma cell invasion via targeting c-Met. Kaohsiung J Med Sci 2020;36:81-8. [Crossref] [PubMed]
- Wang DS, Zhang HQ, Zhang B, et al. miR-133 inhibits pituitary tumor cell migration and invasion via down-regulating FOXC1 expression. Genet Mol Res 2016. [Crossref] [PubMed]
- Grzywa TM, Klicka K, Rak B, et al. Lineage-dependent role of miR-410-3p as oncomiR in gonadotroph and corticotroph pituitary adenomas or tumor suppressor miR in somatotroph adenomas via MAPK, PTEN/AKT, and STAT3 signaling pathways. Endocrine 2019;65:646-55. [Crossref] [PubMed]
- Zhang Y, Pusch S, Innes J, et al. Mutant IDH Sensitizes Gliomas to Endoplasmic Reticulum Stress and Triggers Apoptosis via miR-183-Mediated Inhibition of Semaphorin 3E. Cancer Res 2019;79:4994-5007. [Crossref] [PubMed]
- Fan H, Yuan R, Cheng S, et al. Overexpressed miR-183 promoted glioblastoma radioresistance via down-regulating LRIG1. Biomed Pharmacother 2018;97:1554-63. [Crossref] [PubMed]
- Calsina B, Castro-Vega LJ, Torres-Pérez R, et al. Integrative multi-omics analysis identifies a prognostic miRNA signature and a targetable miR-21-3p/TSC2/mTOR axis in metastatic pheochromocytoma/paraganglioma. Theranostics 2019;9:4946-58. [Crossref] [PubMed]
- Torrejón B, Cristóbal I, Rojo F, et al. Caveolin-1 is Markedly Downregulated in Patients with Early-Stage Colorectal Cancer. World J Surg 2017;41:2625-30. [Crossref] [PubMed]
- Kowalska K, Nowakowska M, Domińska K, et al. Coexpression of CAV-1, AT1-R and FOXM1 in prostate and breast cancer and normal cell lines and their influence on metastatic properties. Acta Biochim Pol 2016;63:493-9. [Crossref] [PubMed]
- Zou W, Ma X, Hua W, et al. Caveolin-1 mediates chemoresistance in cisplatin-resistant ovarian cancer cells by targeting apoptosis through the Notch-1/Akt/NF-κB pathway. Oncol Rep 2015;34:3256-63. [Crossref] [PubMed]
- Raja SA, Shah STA, Tariq A, et al. Caveolin-1 and dynamin-2 overexpression is associated with the progression of bladder cancer. Oncol Lett 2019;18:219-26. [Crossref] [PubMed]
- Shao S, Qin T, Qian W, et al. Cav-1 Ablation in Pancreatic Stellate Cells Promotes Pancreatic Cancer Growth through Nrf2-Induced shh Signaling. Oxid Med Cell Longev 2020;2020:1868764. [Crossref] [PubMed]
- Lin CJ, Yun EJ, Lo UG, et al. The paracrine induction of prostate cancer progression by caveolin-1. Cell Death Dis 2019;10:834. [Crossref] [PubMed]
- Liu W, Yin NC, Liu H, et al. Cav-1 promote lung cancer cell proliferation and invasion through lncRNA HOTAIR. Gene 2018;641:335-40. [Crossref] [PubMed]
- Cao D, Di M, Liang J, et al. MicroRNA-183 in Cancer Progression. J Cancer 2020;11:1315-24. [Crossref] [PubMed]
- Qin S, Shi X, Wang C, et al. Transcription Factor and miRNA Interplays Can Manifest the Survival of ccRCC Patients. Cancers (Basel) 2019;11:1668. [Crossref] [PubMed]
- Zhu Y, Zhu MX, Zhang XD, et al. SMYD3 stimulates EZR and LOXL2 transcription to enhance proliferation, migration, and invasion in esophageal squamous cell carcinoma. Hum Pathol 2016;52:153-63. [Crossref] [PubMed]
- Chen Y, Chuan HL, Yu SY, et al. A Novel Invasive-Related Biomarker in Three Subtypes of Nonfunctioning Pituitary Adenomas. World Neurosurg 2017;100:514-21. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


