
Signaling pathways of genetic variants and miRNAs in the pathogenesis of myasthenia gravis
Introduction
Myasthenia gravis (MG) is a chronic autoimmune neuromuscular disorder causing muscle weakness and characterized by a defect in synaptic transmission at the neuromuscular junction. Currently, several autoimmune antibodies to acetylcholine receptors (AChR), muscle-specific kinase (MuSK), and low molecular weight receptor-related low-density lipoprotein-4 (Lrp4) have been demonstrated to attack the corresponding antigenic targets, leading to the onset of MG (1). However, the involved genetic and molecular mechanisms leading to the induction and production of these antibodies remain unclear.
A variety of genetic variants, e.g., −3279 and IVS9+459 in Foxp3, have been shown to be strongly associated with MG risk (2). Mutations in different genes encoding molecules important in the neuromuscular junction cause major changes in function (3). Nevertheless, several miRNAs, e.g., miR-122 and miR-185 (4) have been reported to be differentially expressed in the serum or peripheral blood mononuclear cells (PBMC) of MG patients, showing the close connections between these miRNAs and the pathophysiology of MG (5). Aberrant microRNA (miRNA) expression suggests that epigenetic modification influences MG risk (4).
Genes are the core of genetics and epigenetics. The overwhelming majority of associated genes are involved in the immune system. Therefore, we presumed that some key genes targeted by these genetic variants or miRNAs, i.e., at the DNA or RNA level, are involved in some critical pathways to trigger the onset of MG. By using bioinformatics tools, we aimed to predict these key genes and signaling pathways. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/gs-20-39).
Methods
A summary of the following steps is shown in Figure 1. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
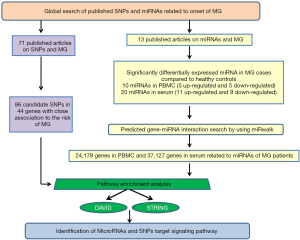
Global search of SNPs and miRNAs in literature databases
To identify publications on MG and genetic variants, a comprehensive, systematic search of existing literature was first conducted. We searched the databases Medline, PubMed and Embase, up to June 1st 2019 using the MeSH terms “Myasthenia Gravis” and “Polymorphism, Single Nucleotide (SNP)”, or “Genome-Wide Association Study (GWAS)”. We performed another search of the literature on MG and miRNAs, using “Myasthenia Gravis” and “miRNAs” as MeSH terms. The search strategies used are listed in Table S1. All the identified publications were dealing with blood samples in MG cases and controls. We excluded articles without full text or not in English. Two studies collected data from these articles (KQ and YD, with 7 years and 12 years of experience in MG, respectively. YD has two years of statistical work experience. Both are familiar with English).
Prediction of miRNA target genes
Target genes of these miRNAs were predicted using the bioinformatics prediction tool “miRWalk” (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/) (6) and validated by all the other tools provided on the miRWalk website including miRanda, miRDB, RNA22, and Targetscan.
Pathway enrichment analyses
In order to identify pathways involving the genes targeted by SNPs or miRNAs, we performed enrichment analysis using online functional annotation tools, i.e., DAVID (http://david.abcc.ncifcrf.gov/, updated in May 2018) (7) and STRING (http://string-db.org/, version 10.0) (8) The top most significant pathways were confirmed by gene counts ≥5, and both satisfied the Bonferroni-corrected cutoff (Bonferroni P value <0.05).
Analysis of protein-protein interaction networks
Protein-protein interaction networks (PPIs) and clusters of these proteins were verified by STRING (confidence scores ≥0.4). The key genes were defined as those with higher degrees of connectivity in PPIs. Cytoscape was utilized to construct the protein interaction network, which was used to calculate the score of gene nodes by using three centrality methods [i.e., Degree Centrality, Betweenness Centrality, and Closeness Centrality (9-11)]. The key genes were defined as those with higher degrees of connectivity in PPIs, which were identified by a network topology analysis (11,12).
Statistical analysis
Statistical analysis was carried out by using the MedCalc Statistical Software version v19.0.3 (MedCalc Software bvba, Ostend, Belgium). The key genes were defined as those with higher degrees of connectivity in PPIs, which were identified by a network topology analysis. The PathExNet tool was also used to isolate the genes that participate in each of the selected pathways based on KEGG 2019 along with the logFC and P value metrics, as provided by the differential expression analysis (13).
Results
We identified 86 candidate SNPs located in 44 related genes from 71 articles (Table S2) and 30 miRNAs from 13 articles (Table S3).
Five genes containing the reported SNPs associated with MG risk were involved in the inflammatory bowel disease (IBD) signaling pathway, and FoxP3 was the key gene
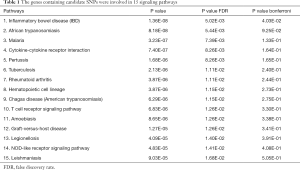
We used DAVID and STRING to identify signaling pathways of the 44 genes containing candidate SNPs (Table S1), and found these genes were involved in 15 signaling pathways (Table 1).
Full table
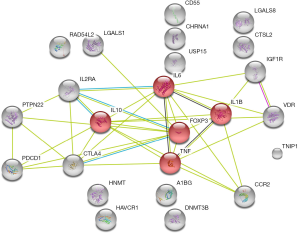
Cytoscape was utilized to construct the protein interaction network, which was used to calculate the score of gene nodes by using three centrality methods [i.e., Degree Centrality, Betweenness Centrality, and Closeness Centrality (9-11)]. Only one significant pathway was identified (gene counts =5, Bonferroni P value <0.040, and confidence scores =0.59), i.e., the IBD signaling pathway, involving five genes (Foxp3, IL6, IL10, IL1B, and TNF). Additionally, in these nodes with high degrees were identified using a network topology analysis, FoxP3 was shown to be the key gene among them (Figure 2, Table 2). This result suggests that the inflammatory and immune may play an important role in the occurrence and development of MG.

Full table
MiRNAs were involved in adherens junction, cancer-related and apoptosis pathways
We identified 24,179 and 37,127 genes as potential targets of the significantly differentially-expressed miRNAs in PBMC and serum, respectively (https://cdn.amegroups.cn/static/public/gs-20-39-01.docx).
In PBMC, 24,179 genes were found to be involved in 28 signaling pathways, but only one highly significant pathway, the adherens junction pathway (FC>0 and FC<0), had a Bonferroni P value <0.05 (Table 3). In serum, 37,127 genes were involved in 36 signaling pathways, and cancer-related pathway (FC<0) and apoptosis pathways (FC>0) seemed to be the most significant pathways among them (Table 3).

Full table
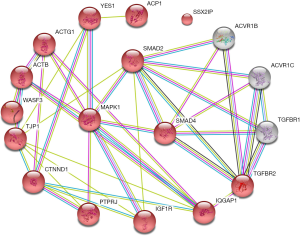
The adherens junction pathway consists of 46 interactions involving 15 genes (Figure 3), key genes including mitogen-activated protein kinase 1 (MAPK1), SMAD family member 4 (SMAD4), and SMAD2 (Figure 3, Table 3).
In serum, the cancer-related pathway involved 67 genes and 379 interactions (Figure 4). However, it was too complicated to visualize these 379 interactions and identify the key genes, probably due to complicated variations and interactions of the genes involved in cancer biology. The apoptosis pathway involved 19 genes and 47 interactions (Figure 5). B-cell lymphoma 2 (BCL2) seemed to be the key gene among them (Figure 5).
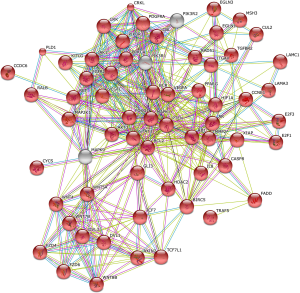
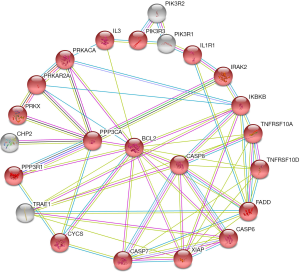
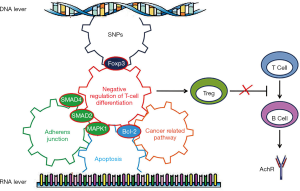
The key genes targeted by SNPs and miRNAs are involved in negative regulation of T cell differentiation
We used STRING to reveal the possible biological processes of five key genes i.e., BCL2, MAPK1, SMAD2, SMAD4 and Foxp3 targeted by miRNAs or SNPs. Intriguingly, we found these key genes seemed to be involved in a pathway which negatively regulates T cell differentiation (Figure 6).
Discussion
We analyzed the mutational gene using bioinformatics, and found that Foxp3 was involved as the key gene in the signaling pathway of IBD, which is a chronic, relapsing inflammatory disorder and an autoimmune disease. Concomitantly, some of the gene mutations found in this study are also mutated in IBD, such as TIM3, IL-10, IL-6, and TNF (14,15). Foxp3 regulates both the development and the function of CD4+CD25+ regulatory T cells (Tregs) (16). Tregs have been proven to control a variety of immune responses to maintain immune homeostasis, ranging from autoimmune diseases to inflammatory conditions (17).
In patients with MG, both the quantities and the functions of Tregs are significantly decreased, suggesting an important role of Tregs in the pathogenesis of MG (17). Tregs in MG patients show decreased expression of Foxp3 and IL-10 indicating a functional deficit. In patients with Foxp3 mutations, Tregs are absent or dysfunctional, always leading to severe autoimmune diseases, e.g., MG and IBD (18). Reduced Treg suppressive activity in MG patients is accompanied by elevated inflammatory cytokines (IL-6, IL-17, TNF-α, and IL-1β), most of which are normally suppressed by Tregs (19). Immunomagnetically-purified Tregs from MG patients were found to suppress the proliferative response of other T cells (20). Leading to imbalance of T/B cells and subsequently affecting the production of auto antibodies (21,22). Additionally, differentiation of Tregs is anticipated to be associated with myasthenia predisposition (23). Overall, the mechanisms of the IBD pathway in MG are unclear, but we speculate that at the DNA level, mutations of Foxp3 apply to Tregs, leading to severe autoimmune diseases via the IBD signaling pathway.
Recent study reveals that imbalance in T follicular helper cells (Tfh) producing IL-17 promotes proinflammatory responses in myasthenia gravis (24). The ratio of Tfh17/Tfh1 has been shown to correlate with a pro-inflammatory and enhanced humoral immune response (25). Preite found that reconstitution of lymphopenic mice with CXCR5-sufficient and CXCR5-deficient Treg cells, as well as non-regulatory memory CD4 T cells, restrained expansion of Tfh and germinal center B cells, and restored germinal center B-cell dynamics and generation of highly mutated, high-affinity antibodies (26). In summary, in the occurrence and development of MG, Tregs control a variety of immune responses to maintain immune homeostasis, ranging from autoimmune diseases to inflammatory conditions (17).
The miRNAs play crucial roles in controlling and modulating immunity (27). Thus far, epidemiology studies have revealed miRNAs differentially expressed in patients with MG (Table S2); however, the target genes and related pathways remain unclear. We used bioinformatics tools to predict target genes and potential pathways of these miRNAs. Our study identified three critical pathways in the onset of MG, including one pathway in PBMC, the adherens junction signaling pathway, and two pathways in serum, the apoptosis and cancer-related pathways.
The adherens junction signal pathway is a key player in the establishment and maintenance of apical-basal cell polarity, regulation of cell proliferation, mobility, and differentiation (28). However, our study found that the differentially-expressed miRNAs in MG may be involved in the adherens junction pathway, in which MAPK1, (also known as Erk2), SMAD 2 and 4 are the key genes. MAPK1 is essential to the signal transduction of extracellular stimuli from the membrane to the nucleus (29). Indeed, the amount of MAPK1 in MG serum was 11.5 times less than in controls (30). In addition, SMADs can activate intracellular TGF-β1 (31). Thereafter, the activated TGF-β1 can induce the generation of CD4+Foxp3+ Tregs (32) and suppress proliferation of AChR-reactive T cells (32). Although the underlying mechanisms of MAPK1, SMAD2, and SMAD4 in the adherens junction pathway during the onset of MG remain unclear, the relationship between this pathway and Tregs warrants further study.
Apoptosis plays an important role via Fas cascades in the onset of many other autoimmune diseases, as well as MG (33). The BCL2 gene located at chromosome 18q21, encodes a 26-kD protein which is an apoptosis inhibitor (34). In thy0517 thymoma cells, BCL2 was found to be overexpressed (35). High expression of BCL2 may cause inhibition of apoptosis in thymocytes, and potentially induce the occurrence of thymoma (36). Although there is no evidence to prove the relationship between MG and BCL2, in another autoimmune disease, systemic lupus erythematosus (SLE), the expression of BCL2 may confer survival and proliferative advantages on Tregs and could represent a possible marker of SLE disease severity (37). Another study revealed that T cell-specific expression of a BCL2 mutant transgene results in enhanced rescue of thymocytes from negative selection, increasing development of Tregs (38). From these findings, we speculate that BCL2 plays an important role in apoptosis signaling through Tregs.
In summary, our study revealed that IBD, adherens junction, apoptosis, and cancer-related signaling pathways are probably involved in the pathogenesis of MG. Intriguingly, all the key genes targeted by SNPs or miRNAs, i.e., Foxp3, SMAD2, MAPK1, SMAD4, and BCL2, seemed to be involved in negative regulation of T cell differentiation. Based on these findings, we hypothesized that SNPs cause the genes to be defective or the miRNAs to down regulate the factors that subsequently negatively regulate Tregs and trigger the onset of MG (Figure 6). Tregs are the core of MG pathogenesis. However, the studies analyzed describe the results from a diverse range of MG cases at different times after onset. The SNPs or miRNAs could also be the results of an immune response to an ongoing insult, immunosuppressive agent or thymus pathology. Therefore, our hypothesis and the underlying mechanisms warrant further robust study.
Acknowledgments
Funding: This study is supported by the Technology Innovation and Application Development Project of Chongqing Province (cstc2019jscx-msxmX0233).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/gs-20-39
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/gs-20-39).The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Muñiz-Castrillo S, Joubert B, Elsensohn MH, et al. Anti-CASPR2 clinical phenotypes correlate with HLA and immunological features. J Neurol Neurosurg Psychiatry 2020;91:1076-84. [Crossref] [PubMed]
- Varade J, Wang N, Lim CK, et al. Novel genetic loci associated HLA-B*08:01 positive myasthenia gravis. J Autoimmun 2018;88:43-9. [Crossref] [PubMed]
- Saruhan-Direskeneli G, Hughes T, Yilmaz V, et al. Genetic heterogeneity within the HLA region in three distinct clinical subgroups of myasthenia gravis. Clin Immunol 2016;166-167:81-8. [Crossref] [PubMed]
- Cavalcante P, Mizrachi T, Barzago C, et al. MicroRNA signature associated with treatment response in myasthenia gravis: A further step towards precision medicine. Pharmacol Res 2019;148:104388. [Crossref] [PubMed]
- Cron MA, Maillard S, Delisle F, et al. Analysis of microRNA expression in the thymus of Myasthenia Gravis patients opens new research avenues. Autoimmun Rev 2018;17:588-600. [Crossref] [PubMed]
- Sticht C, De La Torre C, Parveen A, et al. miRWalk: An online resource for prediction of microRNA binding sites. PLoS One 2018;13:e0206239. [Crossref] [PubMed]
- DiChiacchio L, Sloma MF, Mathews DH. AccessFold: predicting RNA-RNA interactions with consideration for competing self-structure. Bioinformatics 2016;32:1033-9. [Crossref] [PubMed]
- Holt J, McMillan L. Merging of multi-string BWTs with applications. Bioinformatics 2014;30:3524-31. [Crossref] [PubMed]
- Cukierski WJ, Foran DJ. Using Betweenness Centrality to Identify Manifold Shortcuts. Proc IEEE Int Conf Data Min 2008;2008:949-58. [Crossref] [PubMed]
- Du Y, Gao C, Chen X, et al. A new closeness centrality measure via effective distance in complex networks. Chaos 2015;25:033112. [Crossref] [PubMed]
- Gao Y, Zhang S, Zhang Y, et al. Identification of MicroRNA-Target Gene-Transcription Factor Regulatory Networks in Colorectal Adenoma Using Microarray Expression Data. Front Genet 2020;11:463. [Crossref] [PubMed]
- Beg MS, Brenner AJ, Sachdev J, et al. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs 2017;35:180-8. [Crossref] [PubMed]
- Kakouri AC, Votsi C, Tomazou M, et al. Analyzing Gene Expression Profiles from Ataxia and Spasticity Phenotypes to Reveal Spastic Ataxia Related Pathways. Int J Mol Sci 2020;21:E6722. [Crossref] [PubMed]
- Li X, Chen G, Li Y, et al. Involvement of T cell Ig Mucin-3 (Tim-3) in the negative regulation of inflammatory bowel disease. Clin Immunol 2010;134:169-77. [Crossref] [PubMed]
- Suzuki T, Sasahara Y, Kikuchi A, et al. Targeted Sequencing and Immunological Analysis Reveal the Involvement of Primary Immunodeficiency Genes in Pediatric IBD: a Japanese Multicenter Study. J Clin Immunol 2017;37:67-79. [Crossref] [PubMed]
- De Obaldia ME, Bhandoola A. Transcriptional regulation of innate and adaptive lymphocyte lineages. Annu Rev Immunol 2015;33:607-42. [Crossref] [PubMed]
- Zhang Y, Wang HB, Chi LJ, et al. The role of FoxP3+CD4+CD25hi Tregs in the pathogenesis of myasthenia gravis. Immunol Lett 2009;122:52-7. [Crossref] [PubMed]
- van Herk EH, Te Velde AA. Treg subsets in inflammatory bowel disease and colorectal carcinoma: Characteristics, role, and therapeutic targets. J Gastroenterol Hepatol 2016;31:1393-404. [Crossref] [PubMed]
- Thiruppathi M, Rowin J, Ganesh B, et al. Impaired regulatory function in circulating CD4(+)CD25(high)CD127(low/-) T cells in patients with myasthenia gravis. Clin Immunol 2012;145:209-23. [Crossref] [PubMed]
- Truffault F, Nazzal D, Verdier J, et al. Comparative Analysis of Thymic and Blood Treg in Myasthenia Gravis: Thymic Epithelial Cells Contribute to Thymic Immunoregulatory Defects. Front Immunol 2020;11:782. [Crossref] [PubMed]
- Xu H, Zhang M, Li XL, et al. Corrigendum to: "low and high doses of ursolic acid ameliorate experimental autoimmune myasthenia gravis through different pathways" [Journal of Neuroimmunology 281 (2015) 61-67]. J Neuroimmunol 2019;330:181-3. [Crossref] [PubMed]
- Niu L, Jiang J, Yin Y, et al. LncRNA XLOC_003810 modulates thymic Th17/Treg balance in myasthenia gravis with thymoma. Clin Exp Pharmacol Physiol 2020;47:989-96. [Crossref] [PubMed]
- Chen Y, Li S, Huang R, et al. Comprehensive meta-analysis reveals an association of the HLA-DRB1*1602 allele with autoimmune diseases mediated predominantly by autoantibodies. Autoimmun Rev 2020;19:102532. [Crossref] [PubMed]
- Li Y, Guptill JT, Russo MA, et al. Imbalance in T follicular helper cells producing IL-17 promotes pro-inflammatory responses in MuSK antibody positive myasthenia gravis. J Neuroimmunol 2020;345:577279. [Crossref] [PubMed]
- Locci M, Havenar-Daughton C, Landais E, et al. Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity 2013;39:758-69. [Crossref] [PubMed]
- Preite S, Baumjohann D, Foglierini M, et al. Somatic mutations and affinity maturation are impaired by excessive numbers of T follicular helper cells and restored by Treg cells or memory T cells. Eur J Immunol 2015;45:3010-21. [Crossref] [PubMed]
- Runtsch MC, Hu R, Alexander M, et al. MicroRNA-146a constrains multiple parameters of intestinal immunity and increases susceptibility to DSS colitis. Oncotarget 2015;6:28556-72. [Crossref] [PubMed]
- Yang CC, Graves HK, Moya IM, et al. Differential regulation of the Hippo pathway by adherens junctions and apical-basal cell polarity modules. Proc Natl Acad Sci U S A 2015;112:1785-90. [Crossref] [PubMed]
- Chen S, Evans HG, Evans DR. FAM129B/MINERVA, a novel adherens junction-associated protein, suppresses apoptosis in HeLa cells. J Biol Chem 2011;286:10201-9. [Crossref] [PubMed]
- Cheng Z, Qiu S, Jiang L, et al. MiR-320a is downregulated in patients with myasthenia gravis and modulates inflammatory cytokines production by targeting mitogen-activated protein kinase 1. J Clin Immunol 2013;33:567-76. [Crossref] [PubMed]
- Rizzo S, Basso C, Lazzarini E, et al. TGF-beta1 pathway activation and adherens junction molecular pattern in nonsyndromic mitral valve prolapse. Cardiovasc Pathol 2015;24:359-67. [Crossref] [PubMed]
- Chae CS, Kwon HK, Hwang JS, et al. Prophylactic effect of probiotics on the development of experimental autoimmune myasthenia gravis. PLoS One 2012;7:e52119. [Crossref] [PubMed]
- Mai W, Liu X, Fan Y, et al. Up-regulated expression of Fas antigen in peripheral T cell subsets in patients with myasthenia gravis. Clin Invest Med 2012;35:E294. [Crossref] [PubMed]
- Hockenbery D, Nunez G, Milliman C, et al. Bcl-2 is an inner mitochondrial membrane protein that blocks programmed cell death. Nature 1990;348:334-6. [Crossref] [PubMed]
- Wang G, Wang Y, Zhang P, et al. Establishment and characterization of a novel cell line derived from thymoma with myasthenia gravis patients. Thorac Cancer 2015;6:194-201. [Crossref] [PubMed]
- Salakou S, Kardamakis D, Tsamandas AC, et al. Increased Bax/Bcl-2 ratio up-regulates caspase-3 and increases apoptosis in the thymus of patients with myasthenia gravis. In Vivo 2007;21:123-32. [PubMed]
- Goropevšek A, Gorenjak M, Gradisnik S, et al. STAT5 phosphorylation in CD4 T cells from patients with SLE is related to changes in their subsets and follow-up disease severity. J Leukoc Biol 2017;101:1405-18. [Crossref] [PubMed]
- Burger ML, Leung KK, Bennett MJ, et al. T cell-specific inhibition of multiple apoptotic pathways blocks negative selection and causes autoimmunity. Elife 2014;3:e03468. [Crossref] [PubMed]


