
The impact of EGFR gene polymorphisms on the response and toxicity derived from neoadjuvant chemotherapy for breast cancer
Introduction
Neoadjuvant chemotherapy has been used as a standard method for treating locally advanced breast cancer. It can not only improve the success rates of surgical removal and breast conservation, but also provide valuable information about the sensitivity of patients in response to chemotherapy regimens. Pathologic complete response (pCR) rate has been utilized as an effective indicator to evaluate the efficacy of neoadjuvant chemotherapy. To date, the application of neoadjuvant chemotherapy has achieved a considerable rate in several clinical trials, including a preliminary trial of SHPD001 recently accomplished by our department where the participants received a weekly chemotherapy regimen of paclitaxel combined with cisplatin. The results achieved from the trial were promising, with an overall pCR rate reaching 34.4% and a pCR rate reaching 64.7% in triple-negative breast cancer (1). However, pCR could not be achieved in all patients receiving neoadjuvant chemotherapy. Patients who achieved a pCR after neoadjuvant chemotherapy have been shown to exhibit a higher survival rate compared with those without pCR (2). Therefore, in order to facilitate the identification of patients who may respond better to neoadjuvant chemotherapy, the discovery of effective predictive biomarkers is essential.
Epidermal growth factor receptor (EGFR), or human epidermal growth factor receptor 1 (HER1) is a membrane receptor with tyrosine kinase activity that belongs to the epidermal growth factor receptor (HER) family alongside HER2, HER3 and HER4. Playing an essential role in cell proliferation, EGFR has been found to be widely distributed in many malignant tissues such as breast cancer, stomach cancer and lung cancer (3). EGFR overexpression has been shown to promote cell division, leading to an uncontrolled cell growth (4,5). Studies have shown that EGFR overexpression is frequently found in triple-negative breast cancer (6), and is related to poor prognosis (7,8). A study has shown that both the expression level and gene copy number of EGFR were independent adverse prognostic factors for esophageal adenocarcinoma patients treated with neoadjuvant chemotherapy (9). Choura et al. have identified three specific single nucleotide polymorphisms (SNPs) in EGFR gene (rs17337451, rs1140476, rs17290699) that are related to the protein stability of the EGFR dimer structure and hence a potential attribution to the risk of developing breast cancer (10). Moreover, recent studies have shown that certain SNPs are associated with the response and toxicity from chemotherapy in breast cancer (11-14). Chen et al. have reported that fibroblast growth factor receptor (FGFR) 4 rs1966265 and FGFR2 rs2981578 are related to the response and prognosis of breast cancer patients receiving chemotherapy based on docetaxel-epirubicin-cyclophosphamide combinations (11). Therefore, it is theoretically feasible to identify SNPs in the EGFR gene which are associated with the efficacy of neoadjuvant chemotherapy for breast cancer.
Although the EGFR expression has been proven to have a level of close connection with breast cancer, there has been limited research that focused on the association between EGFR gene SNPs and the efficacy and toxicity derived from neoadjuvant chemotherapy for breast cancer. Sobral-Leite et al. have reported a study focused on the evaluation of the EGFR SNP (rs2227983) located in exons in neoadjuvantly treated breast cancer patients from Brazil and Netherlands (15). However, the regimens in their study were complicated including different kinds such as 5-fluorouracil/doxorubicin/cyclophosphamide (FAC), docetaxel, doxorubicin/cyclophosphamide (AC) and paclitaxel/carboplatin., and they did not report the relationship between SNPs and toxicity derived from neoadjuvant chemotherapy. In this study, two SNP sites, rs1468727 and rs845552, located in EGFR intron 13 and intron 19 respectively were selected, which were reported to have clinical significance in the risk of glioma, and in predicting the therapeutic effect and prognosis of glioma (16-20). However, these two SNP sites have not been studied in breast cancer. We hypothesized that these two sites might be associated with the sensitivity to chemotherapy in breast cancer. To verify this hypothesis, we conducted this exploratory analysis.
We present the following article in accordance with the TRIPOD Reporting Checklist. Available at http://dx.doi.org/10.21037/gs-20-330.
Methods
Study population
All patients included in this study participated in either clinical trial SHPD001 (NCT02199418) or SHPD002 (NCT02221999), and were admitted to Renji Hospital, School of Medicine, Shanghai Jiaotong University, between 2013 and 2016. These programs were approved by the ethics committee of Renji Hospital, School of Medicine, Shanghai Jiaotong University and was in strict adherence to the relevant regulations. The trial was conducted in accordance with the Declaration of Helsinki (as revised in 2013). And an informed consent was signed by each participant.
All participants were female patients aged between 18 and 70 years old, who were pathologically diagnosed with primary breast cancer. In order to be included in this study, all the patients were required to undergo pre-treatment hematological examination, and achieve the following results: white blood cell count (WBC) ≥4.0×109/L, neutrophil count (ANC) ≥1.5×109/L, platelet count (PLT) ≥100×109/L, hemoglobin (Hb) ≥90 g/L, AST (SGOT)/ALT (SGPT) ≤1.5 times the upper limit of normal (ULN), creatinine ≤1.5 times ULN, and total bilirubin ≤1.5 times ULN. Patients who were pregnant, had metastatic breast cancer, or had a history of medical conditions such as uncontrolled cardiovascular disease or severe infection that suggested intolerance to neoadjuvant chemotherapy were excluded from the study.
A core needle biopsy from the mass of breast from each patient was taken. Hormone receptor (HR) positive was defined as ER ≥1% or PR ≥1%. HER2 positive was defined as immunohistochemical HER2+++ or FISH amplification according to ASCO/CAP HER2 testing guideline (21).
Neoadjuvant chemotherapy
Each patient in clinical trial SHPD001 and SHPD002 was scheduled to intravenously receive weekly dosage of paclitaxel at 80 mg/m2 for 4 weeks (d1, d8, d15, d22) and weekly dosage of cisplatin 25 mg/m2 for three weeks (d1, d8, d15), in a cycle of every 4 weeks. All patients underwent chemotherapy for four cycles. In addition, trastuzumab was recommended for HER2 positive patients at a weekly basis. HR positive patients in SHPD002, were randomized to receive endocrine therapy or not. Endocrine therapy included aromatase inhibitor for postmenopausal women and gonadotropin releasing hormone agonist for premenopausal counterparts. All patients received mastectomy with axillary lymph node dissection within 2 weeks following neoadjuvant chemotherapy. pCR was defined as the absence of carcinoma in the breast and axillary lymph nodes.
SNP detection
Five milliliters of peripheral blood samples were taken from all participants before neoadjuvant chemotherapy for DNA extraction and SNP genotyping, which were performed by Shanghai Benegene Biotechnology Co., Ltd (Shanghai, China) using MassARRAY system (Sequenom, San Diego, CA, USA). Detailed primer information is shown in Table 1.

Full table
Statistical analysis
Genotype distributions of all patients were assessed for their adherence to the Hardy–Weinberg equilibrium (HWE). Comparisons of genotype and allele frequencies between groups were performed using a two-sided chi-square test. The risk odds ratio (OR) and the 95% confidence interval (CI) were determined by multivariate logistic regression analyses. The test level was P<0.05 and all P values were bilateral. Statistical analysis was performed using statistical software Stata 14.0 (StataCorp LP, College Station, TX, USA).
Results
Basic information of SNPs
Both SNPs were found to be located in the short arm of chromosome 7 within the intron of the EGFR gene (Table 2). Whilst rs1468727 was identified in intron 13 with a major allele C and a minor allele T, rs845552 was found in intron 19 with a major allele A and a minor allele G. Both SNPs were consistent with the HWE law (P>0.05).

Full table
Association between SNPs and characteristics of patients
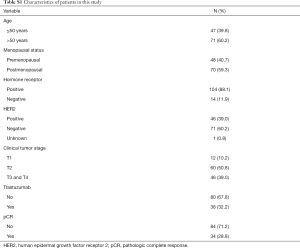
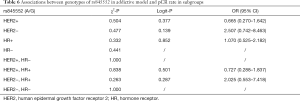
Among all the eligible patients, pretreatment peripheral blood samples were available for 118 patients (Table S1. Our results revealed that rs845552 genotype was related to the HER2 status in co-dominant model and dominant model (Table 3), and that the AA genotype was more prevalent in HER2-positive breast cancer (AA vs. AG, P=0.039; AA vs. GG, P=0.005, in co-dominant model; AA vs. AG+GG, P=0.009, in dominant model). No significant correlation was found between rs1468727 genotype and HER2 status, HR status, Ki67 index and T stage (P>0.05, Table 3).
Full table
Full table
Association between genotype of SNPs and response to neoadjuvant chemotherapy
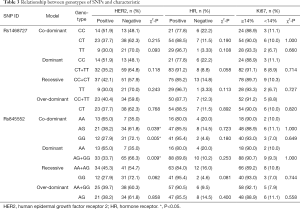
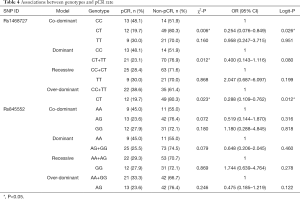
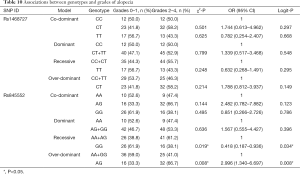
Univariate chi-square analyses showed that different genotypes of rs1468727 in dominant (CC vs. CT+TT, P=0.012), over-dominant (CC+TT vs. CT, P=0.023) and co-dominant models (CC vs. CT, P=0.006) exhibited significant correlation with the efficacy of neoadjuvant chemotherapy (Table 4). Multivariate logistic regression analyses showed that genotypes of rs1468727 were associated with pCR rate in the over-dominant model and the co-dominant model following parameter adjustment in the status of HER2, HR, menopause, and mass size. We showed that patients with a CT genotype were more difficult to achieve pCR compared to patients with a CC+TT genotype (OR =0.288, 95% CI: 0.109–0.762, P=0.012, in over-dominant model, Table 4) or a CC genotype (OR =0.254, 95% CI: 0.076–0.849, P=0.026, in co-dominant model, Table 4). None of the genotypes of rs845552 was found to exhibit a significant association with the pCR rate of neoadjuvant chemotherapy (P>0.05, Table 4).
Full table
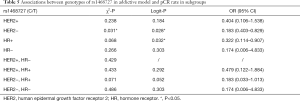
In the HER2-negative subgroup, the rs1468727 genotype was found to be associated with pCR in the addictive model (CC vs. CT vs. TT, P=0.031 in chi-square analyses, and P=0.028 in multivariate logistic regression analyses). Our results showed that pCR was more difficult to be achieved in patients with allele T (OR =0.183, 95% CI: 0.403–0.829, Table 5). Meanwhile, in the HR positive subgroup, multivariate logistic regression analyses showed that the rs1468727 genotype was associated with pCR in the addictive model (CC vs. CT vs. TT, OR =0.322, 95% CI: 0.114–0.907, P=0.032, Table 5). No significant correlation was found between the genotypes of rs845552 and pCR rate in each subgroup (Table 6).
Full table
Full table
Relationship between genotype of SNPs and toxicities of neoadjuvant chemotherapy
We further investigated the association between SNPs and toxicity derived from chemotherapy. Complete information of adverse events was obtained from 109 patients. We found that most patients receiving paclitaxel plus cisplatin developed common adverse events, such as anemia, leukopenia, neutropenia, increased total bilirubin, alopecia, and peripheral neuropathy. Other common grade 3 to grade 4 adverse events included anemia, leukopenia, and neutropenia.
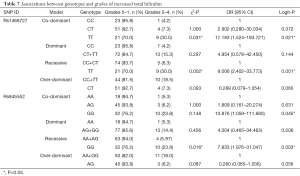
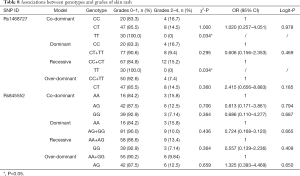
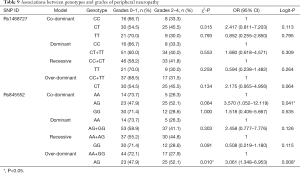
Our analysis revealed that genotypes of the two SNPs were associated with adverse events, including increased total bilirubin, skin rash, peripheral neuropathy, and alopecia (date shown in Tables 7-10). The rs1468727 genotype was associated with increased total bilirubin and skin rash. Multivariate logistic regression analyses revealed that, compared with other genotypes, the TT genotype showed a higher risk of grade 2 to grade 4 increased total bilirubin (CC vs. TT, OR =17.183, 95% CI: 1.524–193.721, P=0.021, in co-dominant model; CC+CT vs. TT, OR =9.006, 95% CI: 2.402–33.773, P=0.001, in recessive model), while univariate chi-square analyses demonstrated a lower risk of grade 2 to grade 4 skin rash (CC vs. TT, P=0.034, in co-dominant model; CC+CT vs. TT, P=0.034, in recessive model). The rs845552 genotype was found to be associated with increased total bilirubin, peripheral neuropathy, and alopecia. The GG genotype was demonstrated through multivariate logistic regression analyses to display a higher risk of grade 2 to grade 4 increased total bilirubin (AA vs. GG, OR =10.876, 95% CI: 1.059–111.680, P=0.045, in co-dominant model; AA+AG vs. GG, OR =7.833, 95% CI: 1.976–31.047, P=0.003, in recessive model), while multivariate logistic regression analyses revealed a lower risk of grade 2 to grade 4 alopecia (AA+AG vs. GG, OR =0.418, 95% CI: 0.187–0.936, P=0.034, in recessive model). Meanwhile, the AG genotype was shown by multivariate logistic regression analyses to exhibit a higher risk of grade 2 to grade 4 peripheral neuropathy (AA vs. AG, OR= 3.570, 95% CI: 1.052–12.119, P=0.041, in co-dominant model; AA+GG vs. AG, OR =3.061, 95% CI: 1.348–6.953, P=0.008, in over-dominant model) and alopecia (AA+GG vs. AG, OR =2.996, 95% CI: 1.340–6.697, P=0.008, in over-dominant model). No obvious association was found between the two SNP genotypes and the risk of anemia, leukopenia and neutropenia (data not shown).
Full table
Full table
Full table
Full table
Discussion
To the best of our knowledge, this is the first report that analyzed EGFR SNPs rs1468727 and rs845552 in breast cancer. In this study, we reported for the first time that the genotype of rs1468727 could predict the treatment response from neoadjuvant chemotherapy for breast cancer. We found that the genotypes of rs1468727 and rs845552 were associated with the degree of neoadjuvant chemotherapy-derived adverse events, such as increased total bilirubin, skin rash, peripheral neuropathy and alopecia. In addition, our results demonstrated for the first time that rs845552 may be related to HER2 status in breast cancer.
We showed that patients with an AA genotype were more likely to be HER2 positive compared to those with a AG or a GG genotype. Rs845552 is known to be located in the intron of EGFR gene. Previous studies have shown that the expression of EGFR can be altered following changes in the intron sequences. The repeat length of CA simple sequence repeat 1 located at the intron 1 of EGFR gene has been reported to be associated with EGFR expression in breast cancer cells (22,23). Consistently, a previous study has reported that both EGFR and HER2 genes tend to be overexpressed simultaneously in cancer patients (24). Thus, the rs845552 genotype may reflect the HER2 status to some extent.
In our study, the rs1468727 genotype was observed to be associated with the treatment response derived from neoadjuvant chemotherapy. In agreement with our results, a previous study has demonstrated a close association between the expression of EGFR protein with the efficacy of neoadjuvant chemotherapy for breast cancer (25). Liu et al. have found that the patients with a high level of EGFR expression can achieve a higher pCR rate while receiving neoadjuvant chemotherapy with docetaxel and epirubicin (26). Tanioka et al. have reported that a high level of EGFR mRNA was associated with the efficacy of neoadjuvant chemotherapy using anthracycline, taxane and trastuzumab (27). Thus, rs1468727, which is a SNP located in the intron region of EGFR, may have an impact on the expression of EGFR at mRNA or protein level, and ultimately impact the sensitivity of neoadjuvant chemotherapy for breast cancer. However, further research is required to verify this hypothesis.
Furthermore, our study suggested that the genotypes of rs1468727 and rs845552 were associated with the degree of adverse events resulting from neoadjuvant chemotherapy, such as increased total bilirubin, skin rash, peripheral neuropathy and alopecia. This could be due to myelin, a protein closely related to the HER family proteins that is localized in the peripheral nervous system (28,29). In agreement with this, a study has reported that EGFR activation is associated with peripheral neuropathy in mice (30). In this study, rs845552 genotype, a SNP of EGFR, was also found to be associated with the occurrence and severity of peripheral neuropathy following chemotherapy, suggesting that EGFR and the peripheral nervous system in patients are closely associated. In addition, we found that genotypes of rs1468727 and rs845552 were associated with the incidence and severity of common adverse events due to chemotherapy, such as increased total bilirubin, skin rash and alopecia, however the specific underlying mechanism remains unclear and demands further studies.
A couple of limitations were identified in our study. Firstly, the sample size in this study was relatively small. However, as an exploratory analysis based on prospective clinical trials, we can get new clues of intrinsic association between these SNPs and the efficacy and toxicity of neoadjuvant chemotherapy for further study. Of course, it is required to expand the sample size to further verify this conclusion in the future. Secondly, the specific mechanism behind the association between the SNPs with the efficacy and toxicity derived from neoadjuvant chemotherapy was unexplored. In future research, we plan to further explore whether and how different SNP genotypes will affect the level of mRNA transcription or protein expression of EGFR, in order to verify its correlation with the efficacy and toxicity of neoadjuvant chemotherapy.
Conclusions
Our study found that, in treating breast cancer with neoadjuvant chemotherapy, EGFR SNP rs1468727 associated with treatment response; while both SNP rs1468727 and rs845552 were attributed to toxicity, such as increased total bilirubin, skin rash, peripheral neuropathy and alopecia. In addition, we also showed that SNP rs845552 may be related to the status of HER2 in breast cancer. Our findings provide important information that may facilitate further studies to dissect the specific mechanism of EGFR in breast cancer.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (81172505, 81302302); the Doctoral Programs Foundation of the Ministry of Education of China (20120071120105); the Shanghai Natural Science Foundation (13ZR1452800, 19ZR1431100); the Shanghai Municipal Commission of Health and Family Planning (20144Y0218, 201640006); the Clinical Research Plan of Shanghai Hospital Development Center (16CR3065B, 12016231); the Shanghai “Rising Stars of Medical Talent” Youth Development Program for Outstanding Youth Medical Talents (2018-16); the Shanghai Collaborative Innovation Center for Translational Medicine (TM201908); the Multidisciplinary Cross Research Foundation of Shanghai Jiaotong University (YG2017QN49, ZH2018QNA42); the Nurturing Fund of Renji Hospital (PYMDT-002); the Science and Technology Commission of Shanghai Municipality (15JC1402700); and the Shanghai Municipal Key Clinical Specialty.
Footnote
Reporting Checklist: The authors have completed the TRIPOD Reporting Checklist. Available at http://dx.doi.org/10.21037/gs-20-330
Data Sharing Statement: Available at http://dx.doi.org/10.21037/gs-20-330
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/gs-20-330). The authors have no conflicts of interest to declare.
Ethics Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards (as revised in 2013). The study was approved by the Institutional Review Board (IRB) of Renji Hospital, School of Medicine, Shanghai Jiaotong University (IRB approval number: [2014]14K and [2017]088). Informed consent was obtained from all individual participants included in the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhou L, Xu S, Yin W, et al. Weekly paclitaxel and cisplatin as neoadjuvant chemotherapy with locally advanced breast cancer: a prospective, single arm, phase II study. Oncotarget 2017;8:79305-14. [Crossref] [PubMed]
- Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet 2014;384:164-72. [Crossref] [PubMed]
- Arteaga CL, Engelman JA. ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell 2014;25:282-303. [Crossref] [PubMed]
- Foley J, Nickerson N, Nam S, et al. EGFR Signaling in Breast Cancer: Bad to the Bone. Semin Cell Dev Biol 2010;21:951-60. [Crossref] [PubMed]
- Macrinici V, Romond E. Clinical updates on EGFR/HER targeted agents in early-stage breast cancer. Clinical Breast Cancer 2010;10:E38-46. [Crossref] [PubMed]
- Rakha EA, El-Sayed ME, Green AR, et al. Prognostic markers in triple-negative breast cancer. Cancer 2007;109:25-32. [Crossref] [PubMed]
- Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 2004;10:5367-74. [Crossref] [PubMed]
- Viale G, Rotmensz N, Maisonneuve P, et al. Invasive ductal carcinoma of the breast with the "triple-negative" phenotype: prognostic implications of EGFR immunoreactivity. Breast Cancer Res Treat 2009;116:317-28. [Crossref] [PubMed]
- Aichler M, Motschmann M, Jutting U, et al. Epidermal growth factor receptor (EGFR) is an independent adverse prognostic factor in esophageal adenocarcinoma patients treated with cisplatin-based neoadjuvant chemotherapy. Oncotarget 2014;5:6620-32. [Crossref] [PubMed]
- Choura M, Frikha F, Kharrat N, et al. Investigating the function of three non-synonymous SNPs in EGFR gene: structural modelling and association with breast cancer. Protein J 2010;29:50-4. [Crossref] [PubMed]
- Chen L, Qi H, Zhang L, et al. Effects of FGFR gene polymorphisms on response and toxicity of cyclophosphamide-epirubicin-docetaxel-based chemotherapy in breast cancer patients. BMC Cancer 2018;18:1038. [Crossref] [PubMed]
- Li XP, Lan JY, Liu DQ, et al. OCA2 rs4778137 polymorphism predicts survival of breast cancer patients receiving neoadjuvant chemotherapy. Gene 2018;651:161-5. [Crossref] [PubMed]
- Wu L, Yao L, Zhang H, et al. A genome-wide association study identifies WT1 variant with better response to 5-fluorouracil, pirarubicin and cyclophosphamide neoadjuvant chemotherapy in breast cancer patients. Oncotarget 2016;7:5042-52. [Crossref] [PubMed]
- Babyshkina N, Zavyalova M, Tarabanovskaya N, et al. Predictive value of vascular endothelial growth factor receptor type 2 in triple-negative breast cancer patients treated with neoadjuvant chemotherapy. Mol Cell Biochem 2018;444:197-206. [Crossref] [PubMed]
- Sobral-Leite M, Lips EH, Vieira-Monteiro HA, et al. Evaluation of the EGFR polymorphism R497K in two cohorts of neoadjuvantly treated breast cancer patients. PLoS One 2017;12:e0189750. [Crossref] [PubMed]
- Yan M, Li J, Na H, et al. A case-control study of the association between the EGFR gene and glioma risk in a Chinese Han population. Oncotarget 2017;8:59823. [Crossref] [PubMed]
- Li B, Zhao W, Li J, et al. Effect of epidermal growth factor receptor gene polymorphisms on prognosis in glioma patients. Oncotarget 2016;7:63054-64. [Crossref] [PubMed]
- Wang X, Zhang H, Wang D, et al. Association of genetic polymorphisms of EGFR with glioma in a Chinese population. Genet Test Mol Biomarkers 2015;19:59-62. [Crossref] [PubMed]
- Yu X, Sun NR, Jang HT, et al. Associations between EGFR gene polymorphisms and susceptibility to glioma: a systematic review and meta-analysis from GWAS and case-control studies. Oncotarget 2017;8:86877-85. [Crossref] [PubMed]
- Huang PH, Xu AM, White FM. Oncogenic EGFR signaling networks in glioma. Sci Signal 2009;2:re6. [Crossref] [PubMed]
- Wolff AC, Hammond MEH, Allison KH, et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J Clin Oncol 2018;36:2105-22. [Crossref] [PubMed]
- Buerger H, Packeisen J, Boecker A, et al. Allelic length of a CA dinucleotide repeat in the egfr gene correlates with the frequency of amplifications of this sequence--first results of an inter-ethnic breast cancer study. J Pathol 2004;203:545-50. [Crossref] [PubMed]
- Tidow N, Boecker A, Schmidt H, et al. Distinct amplification of an untranslated regulatory sequence in the egfr gene contributes to early steps in breast cancer development. Cancer Res 2003;63:1172-8. [PubMed]
- Li JL, Lin SH, Chen HQ, et al. Clinical significance of HER2 and EGFR expression in colorectal cancer patients with ovarian metastasis. BMC Clin Pathol 2019;19:3. [Crossref] [PubMed]
- Singh LC, Chakraborty A, Mishra AK, et al. Study on predictive role of AR and EGFR family genes with response to neoadjuvant chemotherapy in locally advanced breast cancer in Indian women. Med Oncol 2012;29:539-46. [Crossref] [PubMed]
- Liu L, Li XR, Hu YH, et al. Relevance between TOP2A, EGFR gene expression and efficacy of docetaxel plus epirubicin as neoadjuvant chemotherapy in triple negative breast cancer patients. Zhonghua Yi Xue Za Zhi 2016;96:940-3. [PubMed]
- Tanioka M, Sakai K, Sudo T, et al. Transcriptional CCND1 expression as a predictor of poor response to neoadjuvant chemotherapy with trastuzumab in HER2-positive/ER-positive breast cancer. Breast Cancer Res Treat 2014;147:513-25. [Crossref] [PubMed]
- Britsch S, Li L, Kirchhoff S, et al. The ErbB2 and ErbB3 receptors and their ligand, neuregulin-1, are essential for development of the sympathetic nervous system. Genes Dev 1998;12:1825-36. [Crossref] [PubMed]
- Garratt AN, Voiculescu O, Topilko P, et al. A dual role of erbB2 in myelination and in expansion of the schwann cell precursor pool. J Cell Biol 2000;148:1035-46. [Crossref] [PubMed]
- Dahlhoff M, Emrich D, Wolf E, et al. Increased activation of the epidermal growth factor receptor in transgenic mice overexpressing epigen causes peripheral neuropathy. Biochim Biophys Acta 2013;1832:2068-76. [Crossref] [PubMed]


