
Artificial intelligence-assisted HER2 interpretation for breast cancers in a multi-laboratory study
Highlight box
Key findings
• Human epidermal growth factor receptor 2 (HER2) immunohistochemistry (IHC)-1+ and IHC-2+ breast cancers show a low concordance rate in manual interpretation.
• Artificial intelligence (AI)-assisted HER2 interpretation can identify faint/barely perceptible membrane staining, which is an ideal way for HER2-ultralow breast cancers.
What is known and what is new?
• As computational pathology continues to progress, AI-driven approaches are applied to reduce dilemma of HER2 interpretation, such as manual measurement inaccuracies and interobserver variability.
• Disagreements between manual and AI-assisted interpretation occurred significantly more frequently in sections manually scored as 1+ compared to those scored as 0.
What is the implication, and what should change now?
• AI-assisted HER2 interpretation offers a viable approach for multi-laboratory studies, effectively avoiding the subjective errors inherent in manual interpretation.
Introduction
Immunohistochemistry (IHC) for examining the human epidermal growth factor receptor 2 (HER2) protein represents a pivotal step in the diagnosis and treatment of breast cancer. Over the past few decades, the accurate identification of HER2-positive breast cancers, defined as IHC-3+ or IHC-2+ with fluorescence in situ hybridization (FISH) positivity, has been crucial for implementation of HER2-targeted therapies. These targeted treatments lead to improved clinical outcomes for HER2-positive breast cancer patients.
Antibody-drug conjugates (ADCs) have recently emerged as a promising therapy especially in the treatment of HER2-low metastatic breast cancers (1). HER2-low breast cancers are defined as HER2 IHC-1+ or HER2 IHC-2+ with FISH negative. Compared with HER2 IHC-0 or HER2-positive breast cancers, HER2-low breast cancers display unique clinical phenotypes and genetic signatures (2,3). In recognition of these specific features and the associated implications for treatment strategies, Franchet et al. recommended to consider HER2-low breast cancers as a novel entity (4).
However, the routine detection of HER2-low breast cancers is currently reliant on semiquantitative IHC assays. There is a concerning lack of agreement between central and local laboratories in the interpretation of HER2-1+ and IHC-0 cases (5). This inter-laboratory variability may lead to inappropriate treatment decisions for HER2-low breast cancers. With the recent disclosure of DESTINY-Breast06 results (6), a new and crucial challenge has come to the fore: how to accurately distinguish between HER2 zero (where there is no identifiable membrane staining) and HER2-ultralow (HER2 IHC scores falling in the range of >0 to <1+). Many factors, including antibodies, staining procedures, or pathologists’ subjective interpretations, can impact the proportion of HER2-low breast cancers, which currently ranges from 45% to 55% (7).
As computational pathology continues to progress, artificial intelligence (AI)-driven approaches are applied to reduce dilemma of HER2 interpretation, such as manual measurement inaccuracies and interobserver variability. Rule-based models replicate the process of manual interpretation, precisely quantifying the intensity and proportion of the tumor region within tissue slides. One of the key advantages of this type of digital image analysis model avoids the “black-box” conundrum. So it is more palatable and readily acceptable to users. Powered by these models, HER2 interpretation can achieve a high level of interobserver consistency, even when dealing with the particularly challenging HER2-ultralow breast cancers (8,9).
The quality of HER2 IHC slides can significantly impact the accurate interpretation of HER2-low status. In the same case, variations in the cut surfaces of individual slides, along with different experimental conditions, can influence the visibility and apparent expression level of the HER2 protein. As the need to identify breast cancers with faint or minimal HER2 staining becomes more crucial, quality control emerges as an important step for ensuring consistent and reliable HER2 IHC results. With the advantage of AI-assisted HER2 interpretation, which is fast and stable, it may be an ideal tool to conduct real-time monitoring of HER2 testing processes.
In this study, we conducted a multi-institutional investigation to assess the concordance of HER2 examination between manual and AI-assisted HER2 interpretation, with a specific focus on HER2-1+ breast cancers. Additionally, we explored the potential application of AI-assisted HER2 interpretation in routine pathological practice. We present this article in accordance with the TRIPOD reporting checklist (available at https://gs.amegroups.com/article/view/10.21037/gs-2024-560/rc).
Methods
Patient selection
All selected breast cancers were diagnosed between August 2022 and April 2023. Two pathologists reviewed the slides of these cases and defined 2.5-mm-diameter sampling areas of each case in the tissue microarray (TMA). Paraffin blocks of cell line controls (HER2 Four-in-one Pathology Quality Control Product, Suzhou Bogesund Biotechnology Co., Ltd., Suzhou, China) with IHC scores of 0, 1+, 2+, and 3+ were also affixed to each section to perform quality control of the participating laboratories. The first and last sections of the TMA block underwent hematoxylin & eosin (HE) staining and IHC to verify the consistency of sample integrity and HER2 staining intensity. Only tissues with maintained integrity and consistent HER2 staining scores between the first and last sections were included in this study. Ultimately, 53 breast cancer cases from West China Hospital were enrolled in this study. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. The study was approved by ethics committee of West China Hospital (IRS No. 2023-2365). The requirement of informed consent was waived due to the retrospective nature of this study.
Participating laboratories
All laboratories have participated in the regional pathological quality control program every year. At the beginning of this study, pathologists from participating laboratories had received short-term centralism training on HER2 interpretation, guided by the 2018 HER2 guidelines (10), focusing on IHC-0, 1+, and 2+ interpretations.
Completed TMA sections were sent to each participating laboratories within two weeks. All staining (manual or automated) was performed under the standard operating procedure (SOP) of each laboratory and finished within one week of receipt. The HER2 antibodies used in this study were 4B5 (PATHWAY HER2) and MXR001 (Fuzhou MXB Biotechnology Co., Ltd., Fuzhou, China) across different laboratories. Automated IHC staining was conducted on BenchMark System (Roche Diagnostics, North America).
Trained pathologists scored cases (IHC-0 to 3+) through classic microscopes and uploaded results, while central-laboratory experts evaluated staining quality. Results from 35 laboratories passing quality control were included in the analysis.
AI-assisted HER2 interpretation
All the stained TMA sections were returned to the central laboratory. AI-assisted HER2 interpretation was performed using ClinicPath AIM (West China Precision Medicine Industrial Technology Institute), a digital pathology image processing commercial software with a Registration License from the Sichuan FDA Bureau (No. 20232210244). And the images of each case were captured through classic camera-equipped microscope and processed through this software. For each case on every slide, five zones were imaged at 40× magnification. This was done to calculate the percentage of different staining intensities and integrity, and to generate HER2 IHC scores. The illustration of this study is shown in Figure 1.
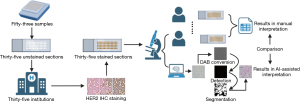
Statistical analysis
Scores from each laboratory were grouped into four (0, 1+, 2+, vs. 3+), three [0, low (1+ and 2+) vs. 3+], or two categories [not 3+ (0, 1+, and 2+) vs. 3+; 0 vs. not 0 (1+, 2+, and 3+); and low (1+ and 2+) vs. not low (0 and 3+)]. Fleiss’ kappa evaluated interrater agreement across laboratories. The intraclass correlation coefficient (ICC) measured within-content relationships for repeated data. The overall percentage agreement (OPA) among laboratories was calculated with the Observers Needed to Evaluate a Subjective Test (ONEST) package (11). A change in the agreement percentage (δ) of no more than 0.005 (0.5%) upon adding an observer was deemed clinical insignificant (12). Kendall’s correlation coefficient between each laboratory was calculated via the corrplot package. A correlation coefficient greater than 0.800 indicated high correlation in this study. For cases with discordant manual interpretations, the Wilcoxon rank-sum test and kappa test assessed agreement between manual and AI-assisted interpretations at each laboratory. Kappa values greater than 0.700 and Wilcoxon P values less than 0.050 were regarded as high agreement. Fleiss’ kappa, ICC, Wilcoxon rank sum test, and kappa test results were calculated using SPSS 26.0 (SPSS, Chicago, IL, USA). The ONEST and corrplot package were utilized in RStudio software version 1.4.1103 (RStudio, Inc., Boston, MA, USA).
Results
Concordance analysis of manual interpretation
In the study, a total of 35 laboratories ultimately participated in the HER2 examinations of 53 breast cancers. Among these laboratories, 14 cases (14/53, 26.4%) exhibited concordant results, including 13 cases with IHC-0 and 1 case with IHC-3+ (Figure 2A). The proportions of cases with IHC-0, IHC-1+, IHC-2+, and IHC-3+ across the participating laboratories were in the ranges of 35.8–72.5%, 9.4–35.8%, 7.5–18.9%, and 3.8–13.2%, respectively (Figure 2B). Analysis based on the Kendall’s correlation coefficient demonstrated that every single one of the 35 laboratories had a high correlation coefficient (greater than 0.800) with at least one other laboratory. Moreover, 65.7% (23/35) of laboratories showed a high correlation coefficient with more than half of the remaining laboratories (Figure S1).
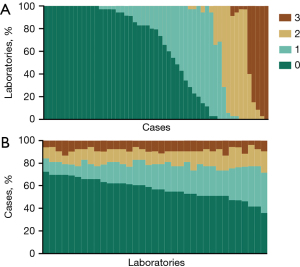
The Fleiss’ kappa value indicated a relatively high degree of concordance in differentiating between cases with an IHC score of 3+ and those with non-3+ cases [kappa =0.866, 95% confidence interval (CI): 0.865–0.866, Table 1]. Conversely, cases with an IHC score of either 1+ or 2+ in at least one laboratory presented with a low OPA and a low Fleiss kappa value (Table 1 and Figure S2).
Table 1
| Categories | Overall percentage agreement (95% CI) | Fleiss’ kappa (95% CI) | ICC (95% CI) |
|---|---|---|---|
| 0, 1+, 2+ vs. 3+ | 0.251 (0.226–0.283) | 0.649 (0.649–0.649) | 0.884 (0.838–0.925) |
| 0, low (1+ and 2+) vs. 3+ | 0.288 (0.283–0.302) | 0.688 (0.687–0.688) | 0.844 (0.785–0.898) |
| Not 3+ (0, 1+, and 2+) vs. 3+ | 0.768 (0.736–0.830) | 0.866 (0.865–0.866) | 0.868 (0.817–0.914) |
| 0 vs. not 0 (1+, 2+, and 3+) | 0.387 (0.377–0.415) | 0.692 (0.692–0.693) | 0.697 (0.607–0.789) |
| Low (1+ and 2+) vs. not low (0 and 3+) | 0.298 (0.283–0.321) | 0.625 (0.624–0.625) | 0.631 (0.534–0.735) |
| Cases which are at least one laboratory scored as | |||
| 0 | 0.301 (0.268–0.341) | 0.495 (0.495–0.495) | 0.557 (0.445–0.687) |
| 1+ | 0.039 (0–0.121) | 0.488 (0.487–0.488) | 0.689 (0.574–0.807) |
| 2+ | 0.024 (0–0.125) | 0.571 (0.570–0.571) | 0.796 (0.662–0.915) |
| 3+ | 0.117 (0.111–0.222) | 0.679 (0.678–0.680) | 0.725 (0.497–0.942) |
CI, confidential interval; ICC, intraclass correlation efficient.
Consistency between manual and AI-assisted interpretation
To assess the disparity between manual and AI-assisted HER2 interpretation, we performed AI-assisted HER2 interpretation on cases lacking manual consistency. Among the 39 cases with non-concordant manual interpretations, 14 (14/39, 35.9%) cases achieved full agreement through AI-assisted HER2 interpretation.
In cases where manual interpretation presented discordant scores restricted to 0 and 1+, 69.6% (16/23) of these cases maintained scores between 0 and 1+ in AI-assisted interpretation. A comparison of all sections from these 23 cases indicated that the percentage of faint/barely perceptible membrane staining in sections shifting from 1+ to 0 (mean ± standard deviation: 3.7%±2.7%) was significantly lower than that in the sections changing from 0 to 1+ (mean ± standard deviation: 13.0%±4.2%, P<0.001, Figure 3A). To determine the cutoff value for the percentage of staining cells between 0 and 1+ in manual interpretation, the specificity for 1+ exceeded 90.00% when the percentage was above 5.4% (Figure 3B). Disagreement between manual and AI-assisted interpretation was more frequent in sections manually scored as 1+ than those scored as 0 (58.6% vs. 2.1%, P<0.001).
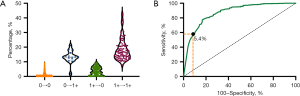
Regarding cases with manual interpretation discordant scores limited to 2+ and 3+, 3 cases (3/6, 50.0%) reached full agreement at 3+ and 2 cases (2/6, 33.3%) achieved full agreement at 2+ via AI-assisted interpretation. Among sections manually scored as 3+, 2 (2/117, 1.7%) shifted to 2+ in AI-assisted interpretation. For sections manually scored as 2+, 20 (20/90, 22.2%) changed scores to 3+ via AI-assisted interpretation.
For cases with manual interpretation discordant scores confined to 1+ and 2+, only 1 case (1/3, 33.3%) reached agreement at 2+ through AI-assisted interpretation. Among slides manually scored as 2+, 6 (6/69, 8.7%) changed to 1+ in AI-assisted interpretation. Among slides manually scored as 1+, 10 (10/36, 27.8%) had scores that changed to 2+ in AI-assisted interpretation.
Influence of different HER2 antibodies on consistency
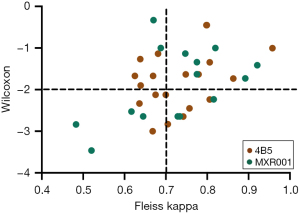
Of the 35 participating laboratories, 20 laboratories (20/35, 57.1%) used 4B5 for HER2 IHC staining, while the remaining 15 laboratories (15/35, 42.9%) utilized MXR001. When comparing the agreement rates between manual and AI-assisted interpretations for 39 cases with discordant manual interpretations across 35 laboratories (Figure 4), the proportion of laboratories achieving high Fleiss kappa values (>0.700) did not differ significantly between those using 4B5 and MXR001 antibodies. Similarly, the significance levels of the Wilcoxon rank sum test showed no significant differences between laboratories using 4B5 and MXR001.
Discussion
In this multi-laboratory study, we investigated the concordance of HER2 examination between manual and AI-assisted HER2 interpretation. Each participating laboratory demonstrated a high correlation coefficient with at least one other laboratory. While all laboratories showed relatively good concordance in distinguishing 3+ cases from non-3+ cases, manual interpretation of HER2-low cases exhibited poor agreement. Notably, AI-assisted interpretation led to complete agreement in nearly one-third of cases with discordant manual interpretations, suggesting that the staining patterns of these cases’ slides may not differ substantially.
The intensity and membrane integrity of tumor cells are key factors in HER2 interpretation. Several studies have utilized machine learning and/or deep learning methods to segment tumor and normal epithelial cells and assign scores to stained tumor cells (13-15). The high precision of these automated scoring systems holds great promise for future applications. Wu et al. demonstrated that an AI-assisted microscope achieved greater consistency and accuracy in HER2 assessment compared to a conventional microscope (9). In the present study, we applied an AI-assisted microscope integrated with a conventional microscope to score HER2 IHC sections. This advanced system can automatically detect tumor cells and evaluate HER2 staining in individual tumor cells, providing a more objective approach to HER2 assessment. Such an objective method is well-suited for evaluating HER2 interpretation across multiple laboratories for the same cases, enabling the identification of interpretation or staining errors. To mitigate histological heterogeneity across sections, we used TMA of the tested samples to standardize the examined areas. Following manual interpretation, AI-assisted interpretation was performed to re-score HER2 for each section of cases with inconsistent results.
In manual HER2 interpretation, a high concordance rate was observed when differentiating 3+ cases from non-3+ cases. Nevertheless, cases scored as 1+ in at least one laboratory exhibited a relatively lower concordance rate compared to the other three categories. Specifically, among 33 cases scored as IHC-1+ in at least one laboratory, 28 (28/33, 84.8%) were rated as IHC-0 in at least one other laboratory. The presence of >10% of tumor cells with faint/barely perceptible, incomplete membrane staining posed a significant challenge in distinguishing IHC-1+ cells from IHC-0 cells under a microscope (10). Breast cancers characterized by <10% of tumor cells with faint/barely perceptible, incomplete membrane staining are classified as HER2 ultralow breast cancers. This subtype of breast cancer has been reported to possess distinct clinical features compared to HER2-low or HER2-null (no staining) breast cancers (16). The DESTINY-Breast06 trial further demonstrated the efficacy of ADCs in the treatment of HER2 ultralow breast cancers (6). Notably, a HER2-null may represent a technical artifact rather than the complete absence of the HER2 protein on the cell membrane (17). Additionally, a previous study reported that the majority of interpretation errors stemmed from misclassifying HER2 ultralow cases as HER2 1+ cases (9).
In this study, we used the 2018 HER2 guidelines’ definition of HER2 IHC-0: no observable staining or <10% of tumor cells faint/barely perceptible membrane staining (10). However, HER2-ultralow breast cancers were defined as HER2 IHC faint/barely membrane staining between 0 and 10%. Among 23 cases with discordant scores restricted to 0 and 1+ during manual interpretation, discrepancies between manual and AI-assisted interpretations were evident, as all these cases had at least one rater with a score difference between 0 and 1+. AI-assisted interpretation detected HER2-ultralow staining patterns in at least one section of these 23 cases. When analyzing all 801 sections of these 23 cases, 145 ratings changed following AI-assisted interpretation (Figure 3A). Specifically, 130 raters (130/145, 89.7%) shifted from IHC-1+ to IHC-0, while only 15 (15/145, 10.3%) changed from IHC-0 to IHC-1+. This high frequency of changes from IHC-1+ to IHC-0 after AI-assisted interpretation aligns with a previous study, which reported that misclassifying HER2 ultralow cases as HER2 1+ was a common source of error (9). These findings suggest that accurately determining the cutoff value for HER2 IHC-1+ is challenging in manual interpretation. By integrating manual ratings with the percentage of membrane staining from AI-assisted interpretation, we found that a cutoff value of 5.4% for IHC-1+ achieved a high specificity (>90.0%) in manual interpretation (Figure 3B). However, this value represents a technical threshold for manual assessment. The optimal cutoff value for IHC-1+ may require further exploration in relation to the treatment outcomes of patients receiving with ADCs. With the recent disclosure of the DB06 clinical trial results in HER2-ultralow breast cancers (6), these promising data may prompt a shift in the focus of HER2 interpretation, prioritizing the identification of truly non-staining cases.
In the present study, cases scored as IHC-0 in at least one laboratory exhibited a relatively low concordance rate during manual interpretation. This finding suggests that the same case could receive an IHC-0 score in one institution and a non-0 score in another. Consistent with our observations, other studies have also reported a low concordance rate when interpreting the same section (9,18). For the 23 cases where manual interpretation discrepancies were confined to scores of 0 and 1+, sections that were identified as having no staining cells by the AI-assisted microscope were all manually scored as IHC-0. Notably, a HER2 IHC-0 result may be more indicative of a technical artifact rather than the complete absence of the HER2 protein on the cell membrane (17). In the present study, cases scored as IHC-0 in at least one laboratory did not uniformly show the absence of membrane staining across all participating laboratories (Figure 5 and Figure S3), even in cases that were consistently scored as 0 in both manual and AI-assisted interpretation. This implies that these cases might have membrane staining, potentially due to variations in cut surfaces or IHC staining protocols. Currently, there is no definitive gold standard for identifying breast cancers with no HER2 expression. However, techniques such as MammaTyper® reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) (19) or RNAscope (20) could potentially serve as viable options for consistently defining breast cancers with absent HER2 expression across different laboratories.
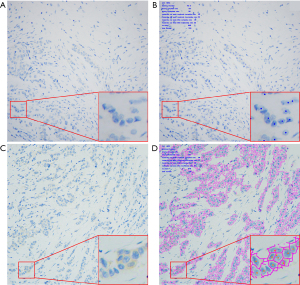
In this study, among the 6 cases with manual interpretation scores restricted to 2+ and 3+, 5 cases (5/6, 83.3%) achieved concordance with AI - assisted interpretation, while the remaining one case (1/6, 16.7%) maintained a score of either 2+ or 3+ with AI-assisted analysis. All 6 cases were confirmed as HER2-positive cases by FISH, indicating that the distinction between IHC scores of 2+ and 3+ has no impact on HER2-targeted therapy. In our prior research on HER2 IHC-2+ breast cancers, FISH-positive cases typically exhibited more intense staining patterns compared to FISH-negative cases (21). The high expression of the HER2 protein in FISH-positive cases rendered them more readily stained, clearly differentiating them from cases with weaker staining.
All participating laboratories used 4B5 or MXR001 antibodies for HER2 IHC staining. There were no significant differences in the proportions of cases with IHC scores of 0 or 1+ between laboratories using 4B5 and those using MXR001 (P=0.59 for IHC-0, P=0.62 for IHC-1+). When analyzing the consistency between manual and AI-assisted interpretations, laboratories using different antibodies showed no significant differences in the rates of high consistency. At the beginning of this study, the center laboratory performed HER2 IHC staining on the first and last sections of the tested TMA to ensure section uniformity across laboratories. The variations observed among laboratories during AI-assisted interpretation highlighted the diversity of staining procedures, even when the same antibody was utilized. These findings suggest that AI-assisted interpretation serve as an optimal approach for conducting internal and external quality control of HER2 IHC staining.
Conclusions
This multi-laboratory study investigated the concordance of HER2 examination in manual and AI-assisted HER2 interpretation, with a particular emphasis on HER2-1+ breast cancers. Our results demonstrate that AI-assisted HER2 interpretation represents a viable alternative for HER2 assessment in multi-laboratory settings, effectively mitigating the subjective errors inherent in manual interpretation.
Acknowledgments
The authors would like to thank Suzhou Bogesund Biotechnology Co., Ltd. (Suzhou, China), for providing the HER2 Four-in-one Pathology Quality Control Product. The authors would also like to thank all the laboratories that participated in this study. Our abstract has been accepted for presentation at the United States and Canadian Academy of Pathology (USCAP) Annual Meeting, Boston, Massachusetts, the United States, between the 22nd and 27th of March, 2025.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://gs.amegroups.com/article/view/10.21037/gs-2024-560/rc
Data Sharing Statement: Available at https://gs.amegroups.com/article/view/10.21037/gs-2024-560/dss
Peer Review File: Available at https://gs.amegroups.com/article/view/10.21037/gs-2024-560/prf
Funding: This study was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://gs.amegroups.com/article/view/10.21037/gs-2024-560/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. The study was granted by ethics committee of West China Hospital (IRS No. 2023-2365). The requirement of informed consent was waived due to the retrospective nature of this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Modi S, Jacot W, Yamashita T, et al. Trastuzumab Deruxtecan in Previously Treated HER2-Low Advanced Breast Cancer. N Engl J Med 2022;387:9-20. [Crossref] [PubMed]
- Jiang C, Perimbeti S, Deng L, et al. Clinical outcomes of de novo metastatic HER2-low breast cancer: a National Cancer Database Analysis. NPJ Breast Cancer 2022;8:135. [Crossref] [PubMed]
- Yang L, Liu Y, Han D, et al. Clinical Genetic Features and Neoadjuvant Chemotherapy Response in HER2-Low Breast Cancers: A Retrospective, Multicenter Cohort Study. Ann Surg Oncol 2023;30:5653-62. [Crossref] [PubMed]
- Franchet C, Djerroudi L, Maran-Gonzalez A, et al. 2021 update of the GEFPICS’ recommendations for HER2 status assessment in invasive breast cancer in France. Ann Pathol 2021;41:507-20. [Crossref] [PubMed]
- Lambein K, Van Bockstal M, Vandemaele L, et al. Distinguishing score 0 from score 1+ in HER2 immunohistochemistry-negative breast cancer: clinical and pathobiological relevance. Am J Clin Pathol 2013;140:561-6. [Crossref] [PubMed]
- Curigliano G, Hu X, Dent RA, et al. Trastuzumab deruxtecan (T-DXd) vs physician’s choice of chemotherapy (TPC) in patients (pts) with hormone receptor-positive (HR+), human epidermal growth factor receptor 2 (HER2)-low or HER2-ultralow metastatic breast cancer (mBC) with prior endocrine therapy (ET): Primary results from DESTINY-Breast06 (DB-06). J Clin Oncol 2024;42:abstr LBA1000.
- Zhang H, Katerji H, Turner BM, et al. HER2-Low Breast Cancers. Am J Clin Pathol 2022;157:328-36. [Crossref] [PubMed]
- Jung M, Song SG, Cho SI, et al. Artificial intelligence-powered human epidermal growth factor receptor 2 (HER2) analyzer in breast cancer as an assistance tool for pathologists to reduce interobserver variation. J Clin Oncol 2022;40:e12543.
- Wu S, Yue M, Zhang J, et al. The Role of Artificial Intelligence in Accurate Interpretation of HER2 Immunohistochemical Scores 0 and 1+ in Breast Cancer. Mod Pathol 2023;36:100054. [Crossref] [PubMed]
- Wolff AC, Hammond MEH, Allison KH, et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J Clin Oncol 2018;36:2105-22. [Crossref] [PubMed]
- Reisenbichler ES, Han G, Bellizzi A, et al. Prospective multi-institutional evaluation of pathologist assessment of PD-L1 assays for patient selection in triple negative breast cancer. Mod Pathol 2020;33:1746-52. [Crossref] [PubMed]
- Han G, Schell MJ, Reisenbichler ES, et al. Determination of the number of observers needed to evaluate a subjective test and its application in two PD-L1 studies. Stat Med 2022;41:1361-75. [Crossref] [PubMed]
- Khameneh FD, Razavi S, Kamasak M. Automated segmentation of cell membranes to evaluate HER2 status in whole slide images using a modified deep learning network. Comput Biol Med 2019;110:164-74. [Crossref] [PubMed]
- Vandenberghe ME, Scott ML, Scorer PW, et al. Relevance of deep learning to facilitate the diagnosis of HER2 status in breast cancer. Sci Rep 2017;7:45938. [Crossref] [PubMed]
- Tewary S, Arun I, Ahmed R, et al. AutoIHC-Analyzer: computer-assisted microscopy for automated membrane extraction/scoring in HER2 molecular markers. J Microsc 2021;281:87-96. [Crossref] [PubMed]
- Chen Z, Jia H, Zhang H, et al. Is HER2 ultra-low breast cancer different from HER2 null or HER2 low breast cancer? A study of 1363 patients. Breast Cancer Res Treat 2023;202:313-23. [Crossref] [PubMed]
- Press MF, Slamon DJ, Flom KJ, et al. Evaluation of HER-2/neu gene amplification and overexpression: comparison of frequently used assay methods in a molecularly characterized cohort of breast cancer specimens. J Clin Oncol 2002;20:3095-105. [Crossref] [PubMed]
- Robbins CJ, Fernandez AI, Han G, et al. Multi-institutional Assessment of Pathologist Scoring HER2 Immunohistochemistry. Mod Pathol 2023;36:100032. [Crossref] [PubMed]
- Badr NM, Zaakouk M, Zhang Q, et al. Concordance between ER, PR, Ki67, and HER2-low expression in breast cancer by MammaTyper RT-qPCR and immunohistochemistry: implications for the practising pathologist. Histopathology 2024;85:437-50. [Crossref] [PubMed]
- Li X, Lee JH, Gao Y, et al. Correlation of HER2 Protein Level With mRNA Level Quantified by RNAscope in Breast Cancer. Mod Pathol 2024;37:100408. [Crossref] [PubMed]
- Yang L, Zhang Z, Li J, et al. A decision tree-based prediction model for fluorescence in situ hybridization HER2 gene status in HER2 immunohistochemistry-2+ breast cancers: a 2538-case multicenter study on consecutive surgical specimens. J Cancer 2018;9:2327-33. [Crossref] [PubMed]


