
Baseline circulating tumor DNA predicts long-term survival outcomes for patients with early breast cancer
Highlight box
Key findings
• Baseline circulating tumor DNA (ctDNA) could potentially predict the long-term survival outcomes for patients with early breast cancer.
What is known and what is new?
• For breast cancer, ctDNA is a potential biomarker not only capable of monitoring the treatment response during neoadjuvant therapy (NAT) or rescue therapy, but also identifying minimal residual disease (MRD) and detecting early relapses after primary treatment.
• In our study, baseline ctDNA was tested in patients with early invasive breast cancer. The results revealed that patients with high-level ctDNA had significantly lower recurrence-free survival (RFS) compared to patients with low-level ctDNA.
What is the implication, and what should change now?
• Our study suggested that detection of ctDNA at diagnosis, before any treatment, could potentially serve as a biomarker to predict the prognosis for patients with early breast cancer. Further follow-up and more studies with large sample sizes are required to confirm these findings.
Introduction
For breast cancer, treatment decision-making and prognosis assessment predominantly rely on the patients’ clinicopathological characteristics. However, patients with the same stage and molecular subtype may still exhibit divergent prognoses even after receiving standard treatment. Although several commercially available gene-based assays, such as Oncotype DX and MammaPrint, have been employed for predicting the risk of recurrence, their applicability is restricted to patients with hormone receptor (HR)-positive/human epidermal growth factor receptor 2 (HER2)-negative status and low tumor burden in axillary lymph nodes (1-3). Therefore, exploring other potential prognostic indicators is of great significance.
Cell-free DNA (cfDNA) refers to fragmented DNA found in the noncellular component of blood, initially reported by Mandel and Metais in 1948 (4). It is thought that cfDNA is released into the bloodstream through apoptosis or necrosis from normal cells and tumor cells (5). In patients with cancer, cfDNA released from tumor cells is referred to as circulating tumor DNA (ctDNA), carrying genetic information of primary tumors and metastatic lesions (6-8). Compared to cfDNA, ctDNA may serve as a more specific indicator in cancer patients to reflect their tumor status.
Various techniques have been applied to detect somatic mutations in ctDNA. Currently, ctDNA sequencing primarily relies on 2 technique platforms: polymerase chain reaction (PCR) and next-generation sequencing (NGS) (9-13). PCR-based methods are highly sensitive and specific, capable of detecting mutant genes even at low mutation frequencies. However, they can only be used for detecting a limited number of hotspot mutations, and cannot be applied to mutation discovery (11,12). In contrast, NGS-based methods offer the advantage of high throughput, enabling the detection of a wider spectrum of mutant genes at a single time point (9,13).
In the field of breast cancer, the value of ctDNA has been extensively investigated for monitoring treatment response during neoadjuvant therapy (NAT) or rescue therapy (14-18), identifying minimal residual disease (MRD) following primary cancer treatment and early detection of disease progression or recurrence based on ctDNA dynamics (16,19-23). Although the prognostic value of ctDNA detected at diagnosis, before any treatment, has been explored in previous studies, there is currently no consensus on whether detection of ctDNA at diagnosis has predictive value for the prognosis of patients with early breast cancer (14-16,19,20,24).
In this study, we investigated the prognostic potential of ctDNA detected at the time of diagnosis in patients with early breast cancer and a NGS panel that encompassed 1,021 cancer-related genes was employed to identify somatic mutations in ctDNA. We present this article in accordance with the REMARK reporting checklist (25) (available at https://gs.amegroups.com/article/view/10.21037/gs-24-115/rc).
Methods
Study design and cohort
This study aimed to investigate whether detection of ctDNA at diagnosis can predict the risk of relapse for patients with early breast cancer. Peripheral blood samples were collected from serial patients scheduled for surgery and healthy women. We extracted cfDNA from plasma of peripheral blood samples. The cfDNA of patients with breast cancer was subjected to deep targeted sequencing and ctDNA was analyzed subsequently. All patients received standard surgical treatment. After completion of surgery, patients received adjuvant therapy according to National Comprehensive Cancer Network (NCCN) guidelines. The association between detection of ctDNA at diagnosis and patient outcomes was analyzed.
Patients with early breast cancer and healthy women were recruited from Peking University People’s Hospital between August 2016 and October 2016. The inclusion criteria were as follows: primary diagnosis of early breast cancer; no metastatic disease at diagnosis; no previous diagnosis of carcinoma. Patients with inoperable tumors and patients who received NAT before primary surgery were excluded. Clinicopathological characteristics of patients were collected, including age, tumor histological type, histological grade, immunohistochemical results, and tumor-node-metastasis (TNM) stage. Follow-up was ended on 15 July 2023. Survival data were available for all patients, with the median follow-up of 81 months. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Peking University People’s Hospital, Beijing (No. 2016PHB210-001). Informed consent was taken from all the patients.
Sample collection and DNA extraction
Blood samples were collected into ethylenediamine tetraacetic acid (EDTA) tubes 1 day before surgery. Peripheral blood lymphocytes (PBLs) and plasma were separated by centrifugation at 1,600 g for 10 minutes. PBLs and plasma were stored at −80 °C until DNA extraction.
Germline DNA from PBLs was extracted by the CWE9600 Blood DNA Kit (Cwbio, Beijing, China). Then, cfDNA was extracted from plasma using an Enhanced Magnetic Circulating DNA Kit (TIANGEN, Beijing, China).
The concentration of cfDNA was quality-controlled and assessed by the Qubit® dsDNA BR Assay Kit (Qubit® dsDNA BR Assay Kit; Thermo Fisher, Waltham, MA, USA). The size distribution of cfDNA was estimated using an Agilent 2100 Bioanalyzer and the DNA HS kit (Agilent Technologies, Santa Clara, CA, USA).
Library preparation
Before library construction, germline DNA from PBLs was sheared into 200–250 bp fragments with the Hieff NGS Ultima DNA Library Prep Kit for MGI (Yeasen, Shanghai, China). More than 20 ng of cfDNA was used for library construction. Unique identifiers (UIDs) were tagged on 200–250 bp cfDNA to distinguish somatic mutations (26). NGS libraries were prepared from cfDNA and germline DNA using the Hieff NGS Ultima DNA Library Prep Kit for MGI (Yeasen). The library concentration was also determined by Qubit® dsDNA BR Assay Kit as mentioned above.
Target region capture and NGS sequencing
Target region enrichment was carried out using a custom-designed panel consisting of 1,021 cancer-related genes (Geneplus-Beijing Co., Ltd., Beijing, China). Which was used to explore the comprehensive genetic properties of breast cancer. After processing the enriched library, NGS sequencing was performed in Geneplus-Beijing Co., Ltd. (CAP/CLIA accredited laboratory) using the Gene+seq 2000 sequencing platform (MGI, Shenzhen, China). About 10 Gb data were generated for cfDNA samples and 1 Gb data for germline DNA.
Data analysis
After filtering the adaptor and low-quality sequence for raw reads, reads were mapped to the reference human genome (hg19) using the Burrows-Wheeler Aligner with default parameters (27,28).
The Genome Analysis Tool Kit (GATK; https:/www.broadinstitute.org/gatk/) and MuTect were used to detect insertions and deletions (indels) and single nucleotide variants (SNVs) (29). The CONTRA (http://contra-cnv.sourceforge.net) was used to identify copy number variants (CNVs) (30). BreakDancer (https://breakdancer.sourceforge.net/) was used to detect tumor-related structural variations (SVs) (31). All mutant genes were manually verified.
Variant allele frequency (VAF) of each mutation was used to calculate ctDNA concentration. The ctDNA concentration was performed as mean tumor molecules (MTM) per milliliter (mL) of plasma, which had been proven reliable in the analysis of ctDNA concentration previously (16,32). The MTM/mL of plasma was calculated by dividing the total number of mutant molecules by the number of targets detected. The MTM per mL calculated as follows: cfDNA extracted (ng) × 1,000 pg × haploid genome equivalent (hGE) × VAF ÷ 1 ng × 3.3 pg per hGE × plasma volume for extraction (mL) (16,32).
Outcomes and statistical analysis
In the study, our objective was to assess whether detection of ctDNA at diagnosis can predict the prognosis for patients with early breast cancer. The recurrence-free survival (RFS) and invasive disease-free survival (iDFS) were analyzed. As STEEP system defined, the iDFS events include ipsilateral invasive breast tumor recurrence, regional invasive breast cancer recurrence, distant recurrence, death attributable to any cause, contralateral invasive breast cancer and second primary invasive cancer (non-breast). RFS events specifically excluded contralateral invasive breast cancer and second primary invasive cancer (non-breast) from iDFS events (33). The RFS and iDFS were defined as the duration from the surgery to the occurrence of the relevant event. The association between detection of ctDNA and patient outcomes were analyzed using the Kaplan-Meier method, and differences were examined using the log-rank test (34). All P values were 2-sided and considered significant at P<0.05. All statistical analyses were performed with the software SPSS 25.0 (IBN Corp., Armonk, NY, USA) and GraphPad Prism (version 9.0; GraphPad Software, San Diego, CA, USA). GraphPad Prism (version 9.0) and R (version 4.3.1; R Foundation for Statistical Computing, Vienna, Austria) were used for graphic drawing.
Results
Characteristics of patients
A total of 90 patients with early breast cancer and 24 healthy women were recruited serially. The 90 patients included 6 patients with ductal carcinoma in situ (DCIS) and 84 patients with invasive breast cancer. All patients were free from distant metastatic disease at the time of diagnosis. The stage of 84 patients with invasive breast cancer were mainly distributed in stage I (n=31, 36.9%) and stage II (n=44, 52.4%). There were only 9 patients with stage III (10.7%). The clinicopathologic characteristics of all 90 patients with breast cancer and 84 patients with invasive breast cancer are displayed in Table 1.
Table 1
| Clinicopathologic characteristics | All patients (n=90) | Invasive breast cancer patients (n=84) |
|---|---|---|
| Age, n (%) | ||
| ≤50 years | 34 (37.8) | 33 (39.3) |
| >50 years | 56 (62.2) | 51 (60.7) |
| Histological type, n (%) | ||
| Invasive ductal carcinoma | 61 (67.8) | 61 (72.6) |
| Specified carcinoma | 23 (25.5) | 23 (27.4) |
| DCIS | 6 (6.7) | – |
| Pathological grade, n (%) | ||
| I | 6 (6.7) | 6 (7.1) |
| II | 48 (53.3) | 48 (57.1) |
| III | 30 (33.3) | 30 (35.7) |
| NA | 6 (6.7) | – |
| LVSI, n (%) | ||
| Negative | 69 (76.7) | 63 (75.0) |
| Positive | 21 (23.3) | 21 (25.0) |
| Tumor stage, n (%) | ||
| Tis | 6 (6.7) | – |
| T1 | 48 (53.3) | 48 (57.1) |
| T2 | 36 (40.0) | 36 (42.9) |
| Lymph node status, n (%) | ||
| Negative | 57 (63.3) | 51 (60.7) |
| Positive | 33 (36.7) | 33 (39.3) |
| Stage, n (%) | ||
| 0 | 6 (6.7) | – |
| I | 31 (34.4) | 31 (36.9) |
| II | 44 (48.9) | 44 (52.4) |
| III | 9 (10.0) | 9 (10.7) |
| ER status, n (%) | ||
| Negative | 25 (27.8) | 21 (25.0) |
| Positive | 65 (72.2) | 63 (75.0) |
| PR status, n (%) | ||
| Negative | 35 (38.9) | 31 (36.9) |
| Positive | 55 (61.1) | 53 (63.1) |
| HER2 status, n (%) | ||
| Negative | 68 (75.6) | 64 (76.2) |
| Positive | 22 (24.4) | 20 (23.8) |
| Ki-67, n (%) | ||
| ≤20% | 54 (60.0) | 48 (57.1) |
| >20% | 36 (40.0) | 36 (42.9) |
| Molecular subtype, n (%) | ||
| HR+/HER2− | 51 (56.7) | 49 (58.3) |
| HR+/HER2+ | 14 (15.6) | 14 (16.7) |
| HR−/HER2+ | 8 (8.9) | 6 (7.1) |
| HR−/HER2− | 17 (18.9) | 15 (17.9) |
NA, not applicable; LVSI, lymph-vascular space invasion; ER, estrogen receptor; PR, progesterone receptor; HR, hormone receptor; HER2, human epidermal growth factor receptor 2; DCIS, ductal carcinoma in situ.
cfDNA concentration
The mean cfDNA concentration of 84 patients with invasive breast cancer was significantly higher than that of healthy women (10.19±5.59 vs. 6.13±2.40 ng/mL, P=0.0003). The cfDNA concentration was higher in patients with invasive breast cancer compared to patients with DCIS, but the difference was not statistically significant (10.19±5.59 vs. 6.96±2.60 ng/mL, P=0.48). No significant difference in cfDNA concentration was observed between DCIS patients and healthy women (6.96±2.60 vs. 6.13±2.40 ng/mL, P>0.99; Figure 1A).
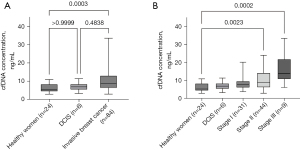
Among patients with invasive breast cancer, the cfDNA concentration showed increasing trend from stage I to stage III. Pairwise comparisons showed that the cfDNA concentration of patients at stage II and stage III were higher than that of healthy women (for stage II, P=0.0023; for stage III, P=0.0002; Figure 1B).
ctDNA detection
Out of 84 patients with invasive breast cancer, 57 patients were identified as ctDNA positive, whereas 27 patients were identified as ctDNA negative based on whether somatic mutation was detected in plasma. The ctDNA detection rate in patients with invasive breast cancer was 67.9% (57/84), with no ctDNA detected in patients with DCIS (P=0.002). As shown in Table 2, among invasive breast cancer patients, there were no significant differences in clinicopathological characteristics between the ctDNA-positive and ctDNA-negative groups.
Table 2
| Clinicopathologic characteristics | Patients (n=84) | ctDNA positive (n=57), n (%) | ctDNA negative (n=27), n (%) | P value |
|---|---|---|---|---|
| Age | 0.15 | |||
| ≤50 years | 33 | 19 (57.6) | 14 (42.4) | |
| >50 years | 51 | 38 (74.5) | 13 (25.5) | |
| Histological type | 0.20 | |||
| Invasive ductal carcinoma | 61 | 44 (72.1) | 17 (27.9) | |
| Specified carcinoma | 23 | 13 (56.5) | 10 (43.5) | |
| Histological grade | 0.34 | |||
| I | 6 | 3 (50.0) | 3 (50.0) | |
| II | 48 | 31 (64.6) | 17 (35.4) | |
| III | 30 | 23 (76.7) | 7 (23.3) | |
| LVSI | 0.79 | |||
| Negative | 63 | 42 (66.7) | 21 (33.3) | |
| Positive | 21 | 15 (71.4) | 6 (28.6) | |
| T stage | 0.49 | |||
| T1 | 48 | 31 (64.6) | 17 (35.4) | |
| T2 | 36 | 26 (72.2) | 10 (27.8) | |
| Lymph node status | 0.82 | |||
| Negative | 51 | 34 (66.7) | 17 (33.3) | |
| Positive | 33 | 23 (69.7) | 10 (30.3) | |
| Stage | 0.56 | |||
| I | 31 | 19 (61.3) | 12 (38.7) | |
| II | 44 | 32 (72.7) | 12 (27.3) | |
| III | 9 | 6 (66.7) | 3 (33.3) | |
| ER status | 0.59 | |||
| Negative | 21 | 13 (61.9) | 8 (38.1) | |
| Positive | 63 | 44 (69.8) | 19 (30.2) | |
| PR status | 0.81 | |||
| Negative | 31 | 22 (71.0) | 9 (29.0) | |
| Positive | 53 | 35 (66.0) | 18 (34.0) | |
| HER2 status | 0.59 | |||
| Negative | 64 | 42 (65.6) | 22 (34.4) | |
| Positive | 20 | 15 (75.0) | 5 (25.0) | |
| Ki-67 | 0.25 | |||
| ≤20% | 48 | 30 (62.5) | 18 (37.5) | |
| >20% | 36 | 27 (75.0) | 9 (25.0) | |
| Molecular subtype | 0.77 | |||
| HR+/HER2− | 49 | 33 (67.3) | 16 (32.7) | |
| HR+/HER2+ | 14 | 11 (78.6) | 3 (21.4) | |
| HR−/HER2+ | 6 | 4 (66.7) | 2 (33.3) | |
| HR−/HER2− | 15 | 9 (60.0) | 6 (40.0) |
ctDNA, circulating tumor DNA; LVSI, lymph-vascular space invasion; ER, estrogen receptor; PR, progesterone receptor; HR, hormone receptor; HER2, human epidermal growth factor receptor 2.
Among the 57 patients with ctDNA positive at diagnosis, a total of 120 single nucleotide polymorphisms (SNPs) in 84 different genes were detected, with 1 to 6 (median, 2) mutations detected in each patient. The mutant genes with higher frequency included TP53 (n=7, 12.3%), TERT (n=4, 7%), FAT1 (n=4, 7%), TSR2 (n=3, 5.3%), SMARCA4 (n=3, 5.3%), NF1 (n=3, 5.3%), and CDH23 (n=3, 5.3%). Additionally, there were 16 genes such as PIK3CA, etc. with mutation frequency of 3.51% and 61 genes with mutation frequency of 1.75% (Figure 2). All mutant genes and corresponding mutation types are displayed in the heatmap in Figure 3.
ctDNA and prognosis
Based on the median concentration of ctDNA (19.6 MTM/mL) among patients with ctDNA detected, we defined ctDNA concentration above 19.6 MTM/mL as high-level ctDNA (n=28) and that below 19.6 MTM/mL or not detected as low-level ctDNA (n=56). The distribution of 84 patients with invasive breast cancer based on the detection and levels of ctDNA at diagnosis, before any treatment, are shown in Figure 4.
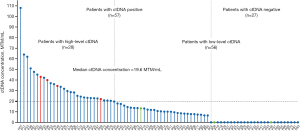
Survival data were available for all patients, with the median follow-up of 81 months (range, 19–83 months). All 4 RFS events occurred in 28 patients with high-level ctDNA, including 2 distant metastases, 1 local recurrence, and 1 ipsilateral axillary lymph node recurrence (displayed in red in Figure 4). Second primary invasive cancers (colorectal cancer, P67 and P46) were detected in 2 patients with low-level ctDNA, and contralateral invasive breast cancer (P08) was detected in 1 patient with low-level ctDNA (labeled in green in Figure 4).
The RFS was significantly lower in patients with high-level ctDNA compared to those with low-level ctDNA (log-rank P=0.0036; HR and 95% CI were unevaluable; Figure 5A). Patients with ctDNA positive had lower RFS than those with ctDNA negative, although there was no statistical difference (log-rank P=0.16; HR and 95% CI were unevaluable; Figure 5B).
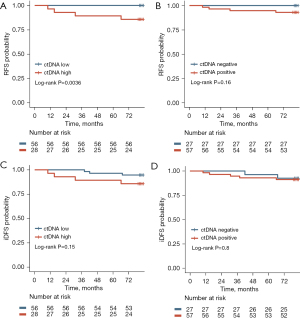
The iDFS of patients with high-level ctDNA was lower compared to that of patients with low-level ctDNA (log-rank P=0.15; HR, 2.87; 95% CI: 0.58–14.09; Figure 5C). Patients with ctDNA positive had lower iDFS than patients with ctDNA negative (log-rank P=0.8; HR, 1.23; 95% CI: 0.25–5.95; Figure 5D), but the differences were not statistically significant.
In subgroup analysis, among patients diagnosed with stage I and stage II disease (75/84=89.3%), only 2 RFS events occurred. The RFS of patients with high-level ctDNA was significantly worse than that of patients with low-level ctDNA (log-rank P=0.037; HR and 95% CI were unevaluable; Figure 6A). Among 63 patients with HR positive, all of the 4 RFS events occurred in patients with high-level ctDNA, whose RFS was significantly lower than that of patients with low-level ctDNA (log-rank P=0.0023; HR and 95% CI were unevaluable; Figure 6B). For 49 patients with HR positive and HER2 negative, RFS was lower in those with high-level ctDNA (log-rank P=0.03; HR and 95% CI were unevaluable; Figure 6C).
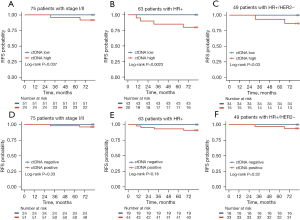
In the aforementioned subgroups, RFS was lower in patients with ctDNA positive compared to those with ctDNA negative, although no statistical difference was observed [for patients diagnosed with stage I and stage II disease, log-rank P=0.33, HR and 95% CI were unevaluable (Figure 6D); for patients with HR positive, log-rank P=0.18, HR and 95% CI were unevaluable (Figure 6E); for patients with HR positive and HER2 negative, log-rank P=0.32, HR and 95% CI were unevaluable (Figure 6F)].
Discussion
The present study suggested that ctDNA at diagnosis, before any treatment, could potentially serve as a biomarker to predict the prognosis for patients with early breast cancer. High-level baseline ctDNA was associated with worse outcomes.
In our study, we observed a significantly higher concentration of cfDNA in patients with invasive breast cancer compared to healthy women. This finding aligned with results of previous studies. Hashad et al. (35) found that the cfDNA concentration in patients with breast cancer was significantly higher than that in patients with benign breast diseases and healthy individuals. Madhavan et al. (36) compared the cfDNA concentrations between 82 patients with early breast cancer and 100 healthy women. The study found that patients with breast cancer had a significantly higher plasma cfDNA concentration than the healthy women.
We used targeted deep sequencing with a NGS panel of 1,021 cancer-related genes to identify somatic mutation in cfDNA from patients with early breast cancer. The large NGS panel enabled the rapid identification and broad coverage of mutant genes, while also ensuring the sensitivity of sequencing. In our study, ctDNA was detected in 67.9% of patients with early breast cancer. This finding was consistent with previous studies, which have reported ctDNA detection rates in patients with early breast cancer at diagnosis ranging from 41% to 72.6% (14-17,19,20,24). Furthermore, the high-frequency mutant genes detected in our study, such as TP53, PIK3CA, and FAT1, were also in line with previous studies (37-39).
Although the prognostic value of ctDNA has been investigated in previous studies, it remains uncertain whether detection of ctDNA at diagnosis has predictive potential for the prognosis of patients with early breast cancer. Magbanua et al. (16) examined the utility of ctDNA for predicting the risk of metastatic recurrence in 84 early breast cancer patients treated in the neoadjuvant I-SPY 2 TRIAL. Their results showed that detection of ctDNA at the time of diagnosis was not associated with an increased risk of metastatic recurrence. Garcia-Murillas et al. (19) used digital PCR (dPCR) to detect ctDNA in a prospective cohort of patients with early breast cancer undergoing NAT. Plasma samples were collected at the time of diagnosis from 42 patients. The study indicated that the detection of ctDNA at diagnosis was not predictive for disease-free survival (DFS) in patients with early breast cancer. However, Garcia-Murillas et al. (20) subsequently conducted another study with larger cohort of 101 patients with early breast cancer receiving NAT, which found that the detection of ctDNA at diagnosis, before any treatment, was associated with RFS. Similarly, Li et al. (14) showed that patients with ctDNA positive at diagnosis had significantly worse outcomes than those with ctDNA negative. In our study, the result showed that patients with ctDNA positive at baseline had a worse prognosis compared to those with ctDNA negative, although the difference was not statistically significant.
These previous studies mentioned above primarily enrolled patients who had relatively higher tumor burden at the time of diagnosis. For instance, in the study conducted by Garcia-Murillas et al. (20), the tumor stage of 101 patients was mainly T2 (n=51, 50.5%) and T3 (n=15, 14.9%), with 51 patients having positive lymph nodes. Similarly, in Li’s study (14), 44 patients mainly presented with tumor stages of T2 (n=23, 52.3%) and T3 (n=14, 31.8%), with the majority having positive lymph nodes (n=35, 80%). In contrast, our study focused on patients at an earlier stage, with all the breast cancer patients at T1 (57.1%) and T2 (42.9%), and only 39.3% of patients having positive lymph nodes. The relatively low number of RFS and iDFS events observed in our study may be attributed to the earlier stage of the patients. This may also explain why the differences we observed in the prognosis between patients with ctDNA positive and ctDNA negative were not statistically significant.
The previous studies have explored the correlation between baseline ctDNA levels and patient prognosis in different types of cancers. Dobilas et al. (40) explored the association of baseline ctDNA levels with prognosis of ovarian cancer patients. They found that the overall survival (OS) of patients with high-level ctDNA was significantly worse than patients with low-level ctDNA. Phallen et al. (41) examined whether baseline ctDNA may be related to disease recurrence and survival of colorectal cancer patients. This study found that patients with increased ctDNA had a shorter progression-free survival (PFS) and OS compared to patients with lower ctDNA level. In our study, patients were categorized into two groups based on baseline ctDNA levels. The result indicated that patients with high-level ctDNA have significantly lower RFS compared to those with low-level ctDNA. It is noteworthy that all RFS events occurred in patients with high-level ctDNA. Taking into account the findings from previous studies (40,41), we believe that exploring the prognostic value of ctDNA at diagnosis in patients with early breast cancer should not only focus on the detection of ctDNA but also on the ctDNA levels.
Despite our study exploring the predictive value of ctDNA in assessing the prognosis of patients with early breast cancer, there were still some limitations. Due to the limited number of RFS events that occurred, we could not perform a multivariate analysis. Longer follow-up is required to achieve conclusions from definite results in the future. In addition, we did not conduct dynamic monitoring for ctDNA after surgery to investigate the relationship between the dynamic changes of ctDNA and patient outcomes.
Conclusions
In conclusion, early breast cancer patients with high-level ctDNA at diagnosis have a worse prognosis. Detection of baseline ctDNA may have the potential to identify high-risk patients. However, further follow-up and more studies with a large sample size are needed.
Acknowledgments
Funding: This study was sponsored by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://gs.amegroups.com/article/view/10.21037/gs-24-115/rc
Data Sharing Statement: Available at https://gs.amegroups.com/article/view/10.21037/gs-24-115/dss
Peer Review File: Available at https://gs.amegroups.com/article/view/10.21037/gs-24-115/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://gs.amegroups.com/article/view/10.21037/gs-24-115/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Peking University People’s Hospital, Beijing (No. 2016PHB210-001). Informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Syed YY, Oncotype DX. Breast Recurrence Score®: A Review of its Use in Early-Stage Breast Cancer. Mol Diagn Ther 2020;24:621-32. [Crossref] [PubMed]
- Brandão M, Pondé N, Piccart-Gebhart M. Mammaprint™: a comprehensive review. Future Oncol 2019;15:207-24. [Crossref] [PubMed]
- Nicolini A, Ferrari P, Duffy MJ. Prognostic and predictive biomarkers in breast cancer: Past, present and future. Semin Cancer Biol 2018;52:56-73. [Crossref] [PubMed]
- Mandel P, Metais P. Nuclear Acids In Human Blood Plasma. C R Seances Soc Biol Fil 1948;142:241-3.
- Corcoran RB, Chabner BA. Application of Cell-free DNA Analysis to Cancer Treatment. N Engl J Med 2018;379:1754-65. [Crossref] [PubMed]
- Pinzani P, D'Argenio V, Del Re M, et al. Updates on liquid biopsy: current trends and future perspectives for clinical application in solid tumors. Clin Chem Lab Med 2021;59:1181-200. [Crossref] [PubMed]
- De Rubis G, Rajeev Krishnan S, Bebawy M. Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends Pharmacol Sci 2019;40:172-86. [Crossref] [PubMed]
- Tzanikou E, Lianidou E. The potential of ctDNA analysis in breast cancer. Crit Rev Clin Lab Sci 2020;57:54-72. [Crossref] [PubMed]
- Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012;4:136ra68. [Crossref] [PubMed]
- Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. [Crossref] [PubMed]
- Gevensleben H, Garcia-Murillas I, Graeser MK, et al. Noninvasive detection of HER2 amplification with plasma DNA digital PCR. Clin Cancer Res 2013;19:3276-84. [Crossref] [PubMed]
- Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011;83:8604-10. [Crossref] [PubMed]
- Wang R, Li X, Zhang H, et al. Cell-free circulating tumor DNA analysis for breast cancer and its clinical utilization as a biomarker. Oncotarget 2017;8:75742-55. [Crossref] [PubMed]
- Li S, Lai H, Liu J, et al. Circulating Tumor DNA Predicts the Response and Prognosis in Patients With Early Breast Cancer Receiving Neoadjuvant Chemotherapy. JCO Precis Oncol 2020;4:PO.19.00292.
- Rothé F, Silva MJ, Venet D, et al. Circulating Tumor DNA in HER2-Amplified Breast Cancer: A Translational Research Substudy of the NeoALTTO Phase III Trial. Clin Cancer Res 2019;25:3581-8. [Crossref] [PubMed]
- Magbanua MJM, Swigart LB, Wu HT, et al. Circulating tumor DNA in neoadjuvant-treated breast cancer reflects response and survival. Ann Oncol 2021;32:229-39. [Crossref] [PubMed]
- Zhou Y, Xu Y, Wang C, et al. Serial circulating tumor DNA identification associated with the efficacy and prognosis of neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat 2021;188:661-73. [Crossref] [PubMed]
- Yi Z, Ma F, Rong G, et al. The molecular tumor burden index as a response evaluation criterion in breast cancer. Signal Transduct Target Ther 2021;6:251. [Crossref] [PubMed]
- Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015;7:302ra133. [Crossref] [PubMed]
- Garcia-Murillas I, Chopra N, Comino-Méndez I, et al. Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol 2019;5:1473-8. [Crossref] [PubMed]
- Turner NC, Swift C, Jenkins B, et al. Results of the c-TRAK TN trial: a clinical trial utilising ctDNA mutation tracking to detect molecular residual disease and trigger intervention in patients with moderate- and high-risk early-stage triple-negative breast cancer. Ann Oncol 2023;34:200-11. [Crossref] [PubMed]
- Stecklein SR, Kimler BF, Yoder R, et al. ctDNA and residual cancer burden are prognostic in triple-negative breast cancer patients with residual disease. NPJ Breast Cancer 2023;9:10. [Crossref] [PubMed]
- Medford AJ, Moy B, Spring LM, et al. Molecular Residual Disease in Breast Cancer: Detection and Therapeutic Interception. Clin Cancer Res 2023;29:4540-8. [Crossref] [PubMed]
- Silva JM, Silva J, Sanchez A, et al. Tumor DNA in plasma at diagnosis of breast cancer patients is a valuable predictor of disease-free survival. Clin Cancer Res 2002;8:3761-6.
- Altman DG, McShane LM, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. BMC Med 2012;10:51. [Crossref] [PubMed]
- Lv X, Zhao M, Yi Y, et al. Detection of Rare Mutations in CtDNA Using Next Generation Sequencing. J Vis Exp 2017;56342. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. [Crossref] [PubMed]
- Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213-9. [Crossref] [PubMed]
- Li J, Lupat R, Amarasinghe KC, et al. CONTRA: copy number analysis for targeted resequencing. Bioinformatics 2012;28:1307-13. [Crossref] [PubMed]
- Chen K, Wallis JW, McLellan MD, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods 2009;6:677-81. [Crossref] [PubMed]
- Magbanua MJM, Brown Swigart L, Ahmed Z, et al. Clinical significance and biology of circulating tumor DNA in high-risk early-stage HER2-negative breast cancer receiving neoadjuvant chemotherapy. Cancer Cell 2023;41:1091-1102.e4. [Crossref] [PubMed]
- Hudis CA, Barlow WE, Costantino JP, et al. Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials: the STEEP system. J Clin Oncol 2007;25:2127-32. [Crossref] [PubMed]
- Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc 1958;53:457-81.
- Hashad D, Sorour A, Ghazal A, et al. Free circulating tumor DNA as a diagnostic marker for breast cancer. J Clin Lab Anal 2012;26:467-72. [Crossref] [PubMed]
- Madhavan D, Wallwiener M, Bents K, et al. Plasma DNA integrity as a biomarker for primary and metastatic breast cancer and potential marker for early diagnosis. Breast Cancer Res Treat 2014;146:163-74. [Crossref] [PubMed]
- Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977;37:646-50.
- Tangvarasittichai O, Jaiwang W, Tangvarasittichai S. The plasma DNA concentration as a potential breast cancer screening marker. Indian J Clin Biochem 2015;30:55-8. [Crossref] [PubMed]
- Zhong XY, Ladewig A, Schmid S, et al. Elevated level of cell-free plasma DNA is associated with breast cancer. Arch Gynecol Obstet 2007;276:327-31. [Crossref] [PubMed]
- Dobilas A, Chen Y, Brueffer C, et al. Preoperative ctDNA Levels Are Associated With Poor Overall Survival in Patients With Ovarian Cancer. Cancer Genomics Proteomics 2023;20:763-70. [Crossref] [PubMed]
- Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017;9:eaan2415. [Crossref] [PubMed]




