
Tumor-derived mesenchymal progenitor cell-related genes in the regulation of breast cancer proliferation
Highlight box
Key findings
• We successfully extracted a specific class of mesenchymal progenitor cells (MPCs) from the cancer tumor microenvironment. We found via biological experiments and gene sequencing that MPCs can influence breast tumor development through specific gene expression. The particular relationship between the tumor microenvironment and tumor tissues was further explored.
What is known and what is new?
• Currently, breast cancer (BC) is the most common malignant tumor in the world. Various studies have investigated the mechanisms of BC development, and recently, an increasing number of researchers have begun to focus on the role of the tumor immune microenvironment, which has been found to play a key role in the development and metastasis of BC.
• We extracted a class of MPCs from the tumor microenvironment, compared their biological properties with those of common MSCs, and sequenced them to identify the differentially expressed genes. Subsequently, the role of tumor-associated mesenchymal progenitor cells differential genes in BC development was examined.
What is the implication, and what should change now?
• In this study, we conducted a preliminary exploration of MPCs and their effect in the breast tumor microenvironment. We briefly characterized the association of differential genes on the proliferation of tumor cells, but additional in-depth studies on the mechanism of these differential genes need to be conducted. Examining the tumor microenvironment can provide new ideas for microenvironment-targeted cancer therapy.
Introduction
Breast cancer (BC) is one of the most common malignant tumors in women. According to the survey data from the International Agency for Research on Cancer (IARC), 2.26 million new cases of BC are reported worldwide each year, and BC has replaced lung cancer as the most common cancer in the world (1). Despite numerous advances being made in BC research in recent years, the etiology of BC remains unclear, and survival rates are hampered by several challenges such as tumor metastasis and drug resistance (2). Therefore, there is an urgent need to identify new effective biomarkers to improve the prognosis and quality of life of patients with BC. The tumor microenvironment (TME) and its components play a key role in regulating tumor drug resistance, recurrence, and metastasis (3). The TME consists of all the nontumor components distributed around tumor cells, including mesenchymal progenitor cells (MPCs), fibroblasts, myeloid-derived suppressor cells (MDSCs), macrophages, lymphocytes, extracellular matrix (ECM), and interwoven vascular endothelial cells (4). Mesenchymal stem cells (MSCs) derived from bone marrow are considered common primitive cells of other stromal cells, and in tumors, MSCs tend to differentiate into tumor stromal precursor cells via the TME, thus becoming tumor-derived MPCs (5). Both MPCs and MSCs have certain stem cell properties; however, MPCs are influenced by tumor cells and the TME and have certain protumorigenic effects compared to MSCs. Research indicates that MPCs are an important component of the TME, can regulate tumor cells, and can, through multiple pathways, promote cancer formation and progression via proliferation, invasive metastasis, immune resistance, and several other malignant biological processes (6,7). Therefore, clarifying the function of MPCs and targeting MPCs can control the occurrence, progression, and metastasis of BC (8,9). RNA sequencing is commonly used to conduct differential expression analysis, ascertain the phenotypic differences between different cells, and to identify particular differential genes.
Stomatin (STOM) is a member of a highly conserved family of intact membrane proteins. Its encoded protein is localized in the cell membrane, where it regulates ion channels and transporter proteins (10). It has been found that STOM is commonly expressed in the nucleus and cytoplasm and is associated with the development of various tumors, such as oral squamous cell carcinoma (11), lung cancer (12), soft tissue sarcoma (13), and pancreatic cancer (14). In addition, the STOM gene is a potential therapeutic target for metastatic BC (15,16). However, few studies have focused on the role of STOM in the microenvironment. Previous findings suggest that the STOM gene is significantly upregulated in the tumor-associated macrophages (TAMs) of colorectal cancer, which may have prognostic and predictive implications for the clinical management of colorectal cancer (17). Laminin subunit alpha 5 (LAMA5) encodes the vertebrate laminin α-chain. It is involved in a variety of biological processes, including cell adhesion, differentiation, migration, signaling, axonal growth, and metastasis (18). LAMA5 is also crucially implicated in the maintenance of the ECM, which is critical for tissue development, stem cell ecological niches, cancer progression, and genetic diseases (19). Collagen and calcium-binding EGF domains 1 (CCBE1) is thought to play a role in ECM remodeling and migration (20). In patients with BC, CCBE1 is frequently downregulated and its absence is associated with reduced recurrence-free survival and overall survival (21). In contrast, high expression levels of CCBE1 in colorectal cancer are associated with high tumor aggressiveness and poor prognosis (22).
Currently, the potential of STOM, CCBE1, and LAMA5 in the diagnosis and pathogenesis of BC microenvironment remains unexamined. Therefore, using transcriptome analysis, we investigated the expression of these genes in BC MPCs and characterize their function in the TME. We present this article in accordance with the MDAR reporting checklist (available at https://gs.amegroups.com/article/view/10.21037/gs-23-387/rc).
Methods
Patient selection and description
This study was performed in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of Chinese People’s Liberation Army (PLA) General Hospital (No. S2016-023-01), and informed consent was taken from all individual participants. In the Breast Surgery Department of the PLA General Hospital, samples of cancerous tissues and adjacent tissues (>5 cm from the tumor) of were obtained from patients with invasive BC who had not received neoadjuvant therapy, with the average volume of the samples being 3 g (Table 1). The pathology results of all samples were confirmed by three specialized pathologists. All samples were stored in saline but not for more than 2 hours so as not to affect the cellular state. The cancer and paracancer samples from the same patient were paired for subsequent experiments.
Table 1
| Patient no. | Sample type | Location | Gender | Age (years) | Pathological type | SBR stage | Pathology subtype | |||
|---|---|---|---|---|---|---|---|---|---|---|
| ER | PR | HER2 | KI67 | |||||||
| 1 | Tumor/adjacent adipose tissues | Right | Female | 47 | Breast invasive ductal carcinoma | III | − | − | 3+ | 45% |
| 2 | Tumor/adjacent adipose tissues | Right | Female | 46 | Breast invasive ductal carcinoma | II | 75% | 80% | 1+ | 60% |
| 3 | Tumor/adjacent adipose tissues | Left | Female | 68 | Breast invasive ductal carcinoma | II | 85% | 90% | 2+ | 25% |
| 4 | Tumor/adjacent adipose tissues | Left | Female | 52 | Breast invasive ductal carcinoma | II | 80% | 85% | 2+ | 20% |
| 5 | Tumor/adjacent adipose tissues | Left | Female | 50 | Breast invasive ductal carcinoma | II | 80% | 80% | 3+ | 40% |
| 6 | Tumor/adjacent adipose tissues | Left | Female | 51 | Breast invasive ductal carcinoma | II | − | − | 0 | 40% |
| 7 | Tumor/adjacent adipose tissues | Right | Female | 44 | Breast invasive ductal carcinoma | III | − | − | 3+ | 45% |
| 8 | Tumor/adjacent adipose tissues | Right | Female | 67 | Breast invasive ductal carcinoma | III | 90% | 90% | 3+ | 85% |
| 9 | Tumor/adjacent adipose tissues | Left | Female | 49 | Breast invasive ductal carcinoma | III | − | − | 2+ | 40% |
| 10 | Tumor/adjacent adipose tissues | Left | Female | 62 | Breast invasive ductal carcinoma | II | 80% | − | 2+ | 35% |
SBR, Scarff-Bloom-Richardson; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
Cell lines
Cell culture of the human triple-negative BC (TNBC) cell lines MDA-MB-231 and MCF-7 were obtained from the American Typical Culture Collection (ATCC). These cell lines have been passed down from the laboratory for about 20 generations or fewer. The MDA-MB-231 and MCF-7 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, Thermo Fisher Scientific, Grand Island, USA), cell cultures were supplemented with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin/streptomycin (Solarbio, Beijing, China), and cells were incubated in a 37 ℃ incubator (with a 5% CO2 atmosphere) for later use.
Isolation and culture of MSCs and MPCs
Fresh BC tissues (n=10) and corresponding normal breast tissues (n=10) were collected from patients with BC. MSCs were obtained from adipose glandular tissues of the normal breast, while MPCs were obtained from BC tissues. Specimens were cut and digested with a calibrated digestion solution consisting of 0.2% collagenase I (Gibco), MSC basic medium (Chemclin Biotech, Beijing, China), and 0.25% trypsin (Biological Industries, Kibbutz Beit-Haemek, Israel) mixed at a ratio of 1:0.4:0.6 at 37 ℃ for 6 h. Cells were then washed with saline solution and resuspended in human MSC special medium (Chemclin Biotech) after centrifugation at 400 × g for 10 min. The cells were seeded in six-well plates at a concentration of 3×106 and incubated at 37 ℃ in a 5% CO2 atmosphere until they reached 80% confluence. Cells were then seeded in T75 culture flasks for expansion and used for subsequent experiments.
Measurement of cell surface markers
We used third-generation cells for surface marker identification. MSCs and MPCs were seeded in six-well plates (1×105 cells/well), and cells were collected when the cell fusion rate reached 80% (1×106 cells). After the cells were washed with phosphate-buffered saline (PBS; with a PH of 7.4), the cells were incubated with 0.5 µL of fluorescein-coupled antibody or 30 min at 4 ℃ under light-proof conditions. The fluorescently labeled antibodies were CD45-FITC, CD14-PE-Cy7, CD73-APC, CD34-PE Cy7, CD105-APC, CD90-FITC, CD146-PE, CD11b-BV605, and CD31-PE (Table 2). Following this, cells were washed using PBS, centrifuged, and resuspended in PBS and then analyzed under a flow cytometer (FACSCelesta, BD Biosciences, San Diego, CA, USA). We adjusted the data for compensation and analyzed the data using FlowJo software (BD Biosciences).
Table 2
| Antibody | Reagent brand | Catalog No. | Lot No. |
|---|---|---|---|
| CD45-FITC | BioLegend | 304006 | B2936670 |
| CD14-PE cy7 | BD Biosciences | 557742 | 70811821 |
| CD73-APC | BioLegend | 344006 | B293700 |
| CD34-PE cy7 | eBioscience | 25-0349-42 | E11311-1633 |
| CD105-APC | BioLegend | 562408 | 6168991 |
| CD90-FITC | BioLegend | 328108 | B304448 |
| CD146-PE | BioLegend | 361006 | B287407 |
| CD11b-BV605 | BD Biosciences | 562721 | 4318545 |
| CD31-PE | eBioscience | 120319-42 | E12826104 |
Cell differentiation assay
After MSCs and MPCs reached 100% confluence, the MSC completion medium was replaced with MSC adipogenic or osteogenic differentiation medium (ScienCell Research Laboratories) and cultured for 20 or 15 days. Adipogenic differentiation and osteogenic differentiation were determined via staining with Oil Red O and Alizarin Red S (Solarbio, China), respectively. Thereafter, observations were made using an optical microscope (Leica, Germany). We used ImageJ software (US National Institutes of Health) for quantitative analysis of cell differentiation.
Cell proliferation assay
MSCs and MPCs grown in the logarithmic phase were washed, digested, and centrifuged, and the cell concentration was adjusted to 1×106 cells/0.5 mL of PBS. Subsequently, 0.5 mL of cell suspension was added with an equal volume of eFluor670 Cell Proliferation Dye (Invitrogen, CA, USA). The mixture was placed at 37 ℃ and cultured in the dark for 10 minutes, after which 5 mL of precooled complete medium was added to terminate the labeling, and the cells were incubated on ice for 5 minutes. The cells were then washed three times with a complete medium, 1×105 cells were resuspended in 200 µL of PBS, and a 4% paraformaldehyde solution of equal volume was added to fix the cell morphology, which was labeled as “0 h”. The remaining number of 1×105 cells was inoculated in a six-well plate. The cells were collected at 24, 48, and 72 h after labeling, and cell fluorescence was measured using a flow cytometer (FACSCalibur, BD Biosciences). Cell proliferation indices were analyzed using ModFit LT 5 (Verity Software House, Topsham, ME, USA).
Cell Counting Kit 8 (CCK8) cell proliferation experiment
MSCs and MPCs were washed three times and then digested and centrifuged, and the cell suspension was prepared. PBS was added to the outer circle of the 96-well plate to prevent the evaporation of the liquid in the plate, with 3,000 cells/100 µL per hole. MSCs and MPCs were seeded into 96-well plates and processed at 0, 48, and 72 h. CCK8 (10 µL) reagent (ScienCell, Carlsbad, CA, USA) was added to each well, and the cell was cultured again for 2 h; the absorbance was measured at 450 nm with a microplate reader, the experimental results were recorded, and a growth curve was drawn.
Scratch assay
Three horizontally spaced horizontal lines were drawn on the back of a six-well plate at equal intervals. Logarithmically grown MSCs and MPCs were collected, washed in PBS, digested, and centrifuged, and cell suspensions were prepared. After counting, the cells were seeded in a six-well plate. The next day, when the cells had fully grown, the cells were scratched with a 200 µL pipette tip in a six-well plate perpendicular to the horizontal line on the back, the original medium was discarded, PBS was applied twice to wash off the cells, and serum-free medium was added. At 0, 6, and 12 h, the cells were positioned according to the horizontal line on the back of the six-well plate and photographed to record the cell invasion.
Transcriptome sequencing of MSCs and MPCs
We selected samples from three patients to extract three sets of paired MSCs and MPCs; specifically, 1×106 of cells from each of the MSC and MPC samples was collected, digested, washed, and frozen in liquid nitrogen. Transcriptome analysis was performed by Annoroad Gene Technology. RNA was extracted and tested for RNA quality; then, messenger RNA (mRNA) was enriched with oligo (dT) and used as a template to synthesize complementary DNA (cDNA). Purification and amplification were applied to duplex DNA, and then cDNA fragments were selected for sequencing with a HiSeq Sequencing System (Illumina, San Diego, CA, USA).
Quantitative real-time polymerase chain reaction (qRT-PCR)
A reviewing the literature for the relevant primers, we designed and synthesized primers We used PrimerBank (https://pga.mgh.harvard.edu/primerbank) to look up the primers and evaluate their quality. Cells were removed from the incubator and washed three times with PBS to extract RNA and test for concentration. RNA was reverse transcribed into cDNA using a TransScript cDNA Synthesis Kit (Transgen, Beijing, China). The cDNA and PerfectStart Green qPCR SuperMix Kit (Transgenics) were then added to a Fluorescent Quantitative PCR Amplification Reaction System (Applied Biosystems, Thermo Fisher Scientific). All primers were obtained from Sangon Biotech, and the primer sequences are listed in Table 3. GAPDH was used as an internal control. Differential gene cycle threshold (Ct) values were calculated via the 2-ΔCt method.
Table 3
| Name | Primer sequence |
|---|---|
| H-GAPDH-F | GAAGGTGAAGGTCGGAGTC |
| H-GAPDH-R | GAAGATGGTGATGGGATTTC |
| H-CCBE1-F | CGACTAAATACCCGTGTCTGAAG |
| H-CCBE1-R | TCGGCACAAACGTCGTAATCT |
| H-STOM-F | GGGAGGGACGCATAGAAGGA |
| H-STOM-R | GTACATTGTTGGAAAGGGAGGC |
| H-LAMA5-F | CCTGGAGAACGGAGAGATCG |
| H-LAMA5-R | CAGCGGCGAGTAGGAGAAAT |
| H-PPARG-F | ACCAAAGTGCAATCAAAGTGGA |
| H-PPARG-R | ATGAGGGAGTTGGAAGGCTCT |
| H-TGF-β-F | CAATTCCTGGCGATACCTCAG |
| H-TGF-β-R | GCACAACTCCGGTGACATCAA |
| H-IL6-F | CCTGAACCTTCCAAAGATGGC |
| H-IL6-R | TTCACCAGGCAAGTCTCCTCA |
| H-IL8-F | ACTGAGAGTGATTGAGAGTGGAC |
| H-IL8-R | AACCCTCTGCACCCAGTTTTC |
| H-CXCL12-F | ATTCTCAACACTCCAAACTGTGC |
| H-CXCL12-R | ACTTTAGCTTCGGGTCAATGC |
| H-IGF-F | GCTCTTCAGTTCGTGTGTGGA |
| H-IGF-R | GCCTCCTTAGATCACAGCTCC |
qRT-PCR, quantitative real-time polymerase chain reaction.
Plasmid transfection
STOM (NM_004099), CCBE1 (NM_133459), and LAMA5 (NM_005560) overexpression plasmids and control plasmids (NewHelix Biotech, Ltd., Shanghai, China) were prepared to transfect MPCs and MSCs using a the Lipofectamine stem cell transfection reagent (Invitrogen, Thermo Fisher Scientific). Transfection was performed by inoculating 1×105 cells per well in 24-well plates. When the cells reached 90% confluence, the transfection mixture was added to the cells, which were then incubated at 37 ℃ for 1–2 days. Following this, the transfected cells were collected for subsequent experiments.
Coculture of BC cells
After culture, MPCs cells were transfected with the target plasmid for 24 h, the transfection efficiency was detected via qRT-PCR, and the cell supernatant was collected and centrifuged at 4 ℃. The conditioned medium (CM) was then obtained by filtrating the cell supernatant using a 0.22-µm filter, and 3,000 cells per well of BC MDA-MB-231 cells were inoculated in a 96-well plate. After cell attachment, the CM was replaced with fresh CM every 24 h. The proliferation of CCK8 cells was measured at 0, 24, 48, and 72 h.
Statistical analysis
Statistical analysis of data was performed using GraphPad Prism version 9.0 analysis software (GraphPad Software, Inc., San Diego, CA, USA). We used the t test to compare two different sample groups, and statistical significance was set at P<0.05. For transcriptome data, we screened for differential genes using |log2 fold change| >1 and P<0.05 and selected appropriate genes with reference to the literature.
Results
Morphology and osteogenic/adipogenic differentiation
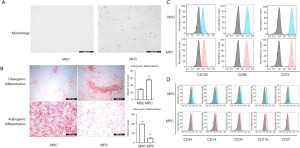
We obtained BC specimens from ten patients and normal tissues specimens at a distance >5 cm from the tumor. MPCs and MSCs isolated from BC tissues and normal breast tissues were both adherent cell with a long spindle-shaped, and some cells were growing in a whirling pattern. There was no significant difference in morphology between MSCs and MPCs when observed microscopically (Figure 1A). Stem cells have the potential to differentiate into multiple cell lineages, such as osteogenic, adipogenic, and chondrogenic types. This property is considered to be an important basis for the identification of stem cells, and to confirm the differentiation potential of our extracted cells, we subjected MSCs and MPCs to differentiation culture. After induction in osteogenic and adipogenic differentiation medium, MSCs and MPCs were able to differentiate successfully. It was found that MPCs were more capable of osteogenic differentiation, while MSCs were more capable of adipogenic differentiation (Figure 1B). We used ImageJ software to quantify the differentiation ability, which corroborated our findings (Figure 1B).
Surface markers of MPCs and MSCs
Stem cells can express a fraction of nonspecific markers, and the Mesenchymal and tissues Stem Cell Committee of the International Society for Cellular Therapy recommends the phenotypic identification of MSCs expressing the CD105, CD73, and CD90 markers and not those expressing CD45, CD34, CD14, CD11b, CD79a, or CD19. To confirm the phenotype, MSCs and MPCs from different patient samples were selected. After incubation, MSCs and MPCs were found to have a phenotype of CD105+, CD73+, CD90+, CD45−, CD14−, CD31−, CD11b−, and CD34− (Figure 1C,1D). By flowcytometric sorting (FACS), we found that both MPCs and MSCs were consistent with stem cell characteristics.
Comparison of the proliferation ability of MSCs and MPCs
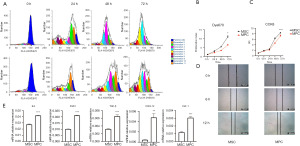
To investigate the difference in proliferation ability of MSCs and MPCs, we used CCK8 assay and eFluor670 cell proliferation assay, and the results of the eFluor670 assay showed that MSCs had stronger proliferation ability than did the MPCs (Figure 2A,2B). Similarly, the results of the CCK8 cell proliferation assay showed that MSCs had a stronger proliferation ability than did the MPCs at 24, 48, and 72 h (Figure 2C).
Migration ability of MSCs and MPCs
To investigate the difference in migration ability between MSCs and MPCs, we implemented a scratch assay The experimental results showed that there was no significant difference in the migratory ability between MPCs and MSCs (Figure 2D).
Comparison of RNA for secreted factors in MSCs and MPCs
We verified the RNA of different secretory factors of MSCs and MPCs using qRT-PCR. MPCs were found to secrete a variety of inflammatory factors including interleukin 6 (IL-6), C-X-C motif chemokine ligand 12 (CXCL12), fibroblast growth factor 2 (FGF2), transforming growth factor β (TGF-β), insulin-like growth factor 1 (IGF-1), and other inflammatory factors (Figure 2E).
Comparison of the transcriptome sequencing of MSCs and MPCs
To determine the differential gene expression between MSCs and MPCs, we clarified the transcriptional changes in the genes of MSCs and MPCs in three patients with BC using transcriptome sequencing. We used transcriptome sequencing for differential expression analysis of the three data sets, and the obtained genes were quantified with an adjusted P value of <0.05 and log2 fold change (FC) >1 used as the differential gene screening threshold (Figure 3A). A total of 156 differential genes were identified between MPCs and MSCs, including 89 upregulated genes and 67 downregulated genes (Figure 3B). CCBE1, LAMA5, and STOM were significantly upregulated, while matrix metallopeptidase 13 (MMP13), IL32, and C-C motif chemokine ligand 11 (CCL11) were significantly downregulated in MPCs (Figure 3C). Gene ontology (GO) analysis showed that for biological process (BP), the differential genes of MPCs and MSCs were mainly enriched in cellular process, biological regulation, and regulation of BP; in molecular function (MF), the differences were mainly in binding and catalytic activity; in cell composition (CC), the differences were mainly in the cell, organelle, and membrane (Figure 3D). The Kyoto Encyclopedia of Genes and Genomes (KEGG) database showed that differential genes were mainly enriched in ECM receptor interactions, PI3K-AKT pathway, adhesive patch-related pathway, cytokine-cytokine receptor interactions, and Hippo pathway (Figure 3E).
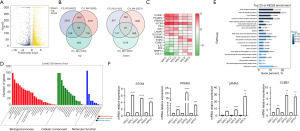
qRT-PCR analysis of the transcriptome pairs of upregulated genes
To validate the transcriptome sequencing of genes, we used qRT-PCR to detect the expression of highly expressed genes including CCBE1, STOM, LAMA5, and peroxisome proliferator-activated receptor gamma (PPARG). At the mRNA level, these genes were upregulated in MPCs (n=3) but not in MSCs (Figure 3F). Because of the heterogeneity across patients, we paired the patient samples (in Figure 3F); for instance, MPC1 was paired with MSC1 and MPC2 was paired with MSC2.
Upregulation of inflammatory factor RNA expression after gene transfection
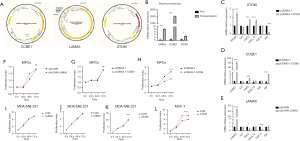
To confirm the effect of STOM, LAMA5, and CCBE1on MPCs, we transfected the STOM-, LAMA5-, and CCBE1-overexpression plasmid and confirmed the gene overexpression via qRT-PCR (Figure 4A,4B). The expression of CXCL12 and FGF2 (Figure 4C) was found to be upregulated in MPCs overexpressing STOM (Figure 4D), IGF was upregulated in MPCs overexpressing CCBE1, and FGF1 (Figure4E) was upregulated in MPCs overexpressing LAMA5.
Effect of the overexpression of target genes on the proliferation of MPCs
To further confirm the effect of the target genes on MPCs, we examined the proliferation of MPCs after transfection with CCK8. The results showed that the STOM gene had an effect on cell proliferation, with overexpression of STOM resulting in a retarded proliferation of MPCs (Figure 4F); meanwhile, CCBE1 (Figure 4G) and LAMA5 (Figure 4H) had no obvious pro-proliferation effect on MPCs.
Effect of MPCs overexpressing the target genes on BC proliferation
We collected supernatants overexpressing the target genes and prepared them as a selective medium for the culture of BC MDA-MB-231 cells. The CCK8 results showed that there was no significant difference between the experimental group and the control group for LAMA5 (Figure 4I) or CCBE1 (Figure 4J), but MPCs overexpressing STOM could promote the proliferation of BC (Figure 4K). We subsequently validated the effect of STOM on MCF-7 BC cell line and found that STOM had the same pro-proliferative effect in noninvasive BC MCF-7 cells (Figure 4L).
Discussion
MPCs are critically involved in the regulation of various types of cells in the tumor and TME (23). New technologies such as single-cell sequencing and spatial transcriptomics have revealed the heterogeneity of MPCs (24,25), and the protumor proliferation and tumor drug resistance properties of MPCs have recently been identified (26-28). However, the details of the mechanisms which primarily influence MPCs to promote breast tumor development still need to be clarified. The discovery of novel targets could inform the development of targeted therapy for BC (29). This study aimed to identify several properties of MPCs and MSCs but, unfortunately, did not explore their effects on BC, and thus the relationship between MPCs and BC should be examined in future research.
In this study, we isolated and extracted MPCs from BC tumors and adjacent healthy tissues; we discovered and both normal breast tissues MSCs and tumor-derived MPCs had similar morphology, while both MSCs and MPCs expressed the same surface markers, including CD105, CD73, CD73, CD54, and CD90. In addition, MSCs and MPCs both demonstrated osteogenic and adipogenic differentiation abilities. However, tumor-derived MPCs exhibited a slower growth rate than did MSCs; consequently, we attempted to explore the biological functional properties of MPCs via transcriptome sequencing. The transcriptome results indicated that MPCs derived from tumor tissues were enriched with 89 upregulated genes and 67 downregulated genes compared to MSCs. The genes highly expressed in BC-derived MPCs included CCBE1, LAMA5, and STOM, which were found to be involved in cell adhesion, ECM receptor interaction, adherent patch-related pathway, and cytokine-cytokine receptor interaction pathways according GO and KEGG pathway analysis. Thus, MPCs may play a key role in the TME and serve as a link between communicating tumor cells.
STOM is a member of the mammalian stomatin-domain protein family, which is named after hereditary human hemolytic anemia (30). The increase in STOM expression has been observed in many cancers. In our study, MPCs overexpressing STOM promoted tumor proliferation to a degree. Moreover, we found that these MPCs could release inflammatory factors, such as CXCL12 and FGF2, at the genetic level. CXCL12 is also referred to as stromal cell-derived factor 1 (SDF1) and plays a key role in tumor development via the CXC chemokine receptor (CXCR4). CXCL12 is significantly associated with invasive metastasis in BC (31), and studies have shown that high CXCL12 expression is a poor prognostic indicator for those with BC (32,33). FGF2, as a member of the FGF family, can promote tumor angiogenesis by acting on FGF receptor (34) and can promote the proliferation and migration of a variety of tumors (35). Moreover, research suggests that FGF2 can promote BC proliferation through ERK signaling (36), yet the mechanism related to the downstream pathway triggered by STOM remains to be explored. We found that STOM overexpression in MPCs slowed the growth of MPCs; in contrast, the overexpression of STOM in MPCs cocultured with BC cell lines resulted in accelerated BC proliferation. We speculate that this phenomenon may be caused by the secretion of pro-tumor proliferation cytokines by MPCs. Additional studies are needed to clarify the specific relationship between MPCs and BC.
CCBE1 and LAMA5 were not found to have a significant role in MPCs. Although they were associated with the release of inflammatory factors including IGF and FGF1, the tumor proliferation effect in vitro was not significantly affected. Nonetheless, CCBE1 plays an important role in lymphangiogenesis and angiogenesis (37), while both the adhesion and angiogenesis effects of LAMA5 may be closely related to tumor metastasis (38,39), which still needs to be investigated further.
Clarifying the genomics of MPCs may help us to better understand the heterogeneity of MPCs and their role in the TME and aid in identifying therapeutic targets. In turn, this can contribute to more efficiently targeting the TME to exert an antitumor effect.
Conclusions
Our study identified certain similarities between BC-derived MPCs and normal breast tissue-derived MSCs, while differences were also observed, chiefly related to the proliferation rate and differentiation ability. Based on transcriptome analysis, we found that the STOM genes can regulate the proliferation of MPCs and BC, but the exact mechanisms underlying this process remain unclear. We also examined the differences between MPCs and MSCs in the TME and determined the distinct characteristics of MPCs, which may provide a reference for subsequent targeted therapies in the BC TME. However, more studies are needed to investigate the related mechanisms and isolate more specific targets within this context.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://gs.amegroups.com/article/view/10.21037/gs-23-387/rc
Data Sharing Statement: Available at https://gs.amegroups.com/article/view/10.21037/gs-23-387/dss
Peer Review File: Available at https://gs.amegroups.com/article/view/10.21037/gs-23-387/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://gs.amegroups.com/article/view/10.21037/gs-23-387/coif). X.L. serves as an Editor-in-Chief of Gland Surgery from May 2022 to April 2024. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of Chinese People’s Liberation Army (PLA) General Hospital (No. S2016-023-01), and informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Hill BS, Sarnella A, D’Avino G, et al. Recruitment of stromal cells into tumor microenvironment promote the metastatic spread of breast cancer. Semin Cancer Biol 2020;60:202-13. [Crossref] [PubMed]
- Chen Y, McAndrews KM, Kalluri R. Clinical and therapeutic relevance of cancer-associated fibroblasts. Nat Rev Clin Oncol 2021;18:792-804. [Crossref] [PubMed]
- Takeuchi S, Yamanouchi K, Sugihara H, et al. Differentiation of skeletal muscle Mesenchymal progenitor cells to myofibroblasts is reversible. Anim Sci J 2020;91:e13368. [Crossref] [PubMed]
- Sarrio D, Franklin CK, Mackay A, et al. Epithelial and mesenchymal subpopulations within normal basal breast cell lines exhibit distinct stem cell/progenitor properties. Stem Cells 2012;30:292-303. [Crossref] [PubMed]
- Sahai E, Astsaturov I, Cukierman E, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer 2020;20:174-86. [Crossref] [PubMed]
- Jackson WM, Nesti LJ, Tuan RS. Potential therapeutic applications of muscle-derived mesenchymal stem and progenitor cells. Expert Opin Biol Ther 2010;10:505-17. [Crossref] [PubMed]
- Sounni NE, Noel A. Targeting the tumor microenvironment for cancer therapy. Clin Chem 2013;59:85-93. [Crossref] [PubMed]
- Klipp RC, Cullinan MM, Bankston JR. Insights into the molecular mechanisms underlying the inhibition of acid-sensing ion channel 3 gating by stomatin. J Gen Physiol 2020;152:e201912471. [Crossref] [PubMed]
- Wang D, Qi H, Li A, et al. Coexisting overexpression of STOML1 and STOML2 proteins may be associated with pathology of oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol 2020;129:591-599.e3. [Crossref] [PubMed]
- Chen JC, Cai HY, Wang Y, et al. Up-regulation of stomatin expression by hypoxia and glucocorticoid stabilizes membrane-associated actin in alveolar epithelial cells. J Cell Mol Med 2013;17:863-72. [Crossref] [PubMed]
- Arkhipova KA, Sheyderman AN, Laktionov KK, et al. Simultaneous expression of flotillin-1, flotillin-2, stomatin and caveolin-1 in non-small cell lung cancer and soft tissue sarcomas. BMC Cancer 2014;14:100. [Crossref] [PubMed]
- Zhu J, Shu X, Guo X, et al. Associations between Genetically Predicted Blood Protein Biomarkers and Pancreatic Cancer Risk. Cancer Epidemiol Biomarkers Prev 2020;29:1501-8. [Crossref] [PubMed]
- Duan J, Bao C, Xie Y, et al. Targeted core-shell nanoparticles for precise CTCF gene insert in treatment of metastatic breast cancer. Bioact Mater 2022;11:1-14. [Crossref] [PubMed]
- Dun MD, Chalkley RJ, Faulkner S, et al. Proteotranscriptomic Profiling of 231-BR Breast Cancer Cells: Identification of Potential Biomarkers and Therapeutic Targets for Brain Metastasis. Mol Cell Proteomics 2015;14:2316-30. [Crossref] [PubMed]
- Cui D, Yuan W, Chen C, et al. Identification of colorectal cancer-associated macrophage biomarkers by integrated bioinformatic analysis. Int J Clin Exp Pathol 2021;14:1-8.
- Sampaolo S, Napolitano F, Tirozzi A, et al. Identification of the first dominant mutation of LAMA5 gene causing a complex multisystem syndrome due to dysfunction of the extracellular matrix. J Med Genet 2017;54:710-20. [Crossref] [PubMed]
- Dey D, Fischer NG, Dragon AH, et al. Culture and characterization of various porcine integumentary-connective tissue-derived mesenchymal stromal cells to facilitate tissue adhesion to percutaneous metal implants. Stem Cell Res Ther 2021;12:604. [Crossref] [PubMed]
- Roukens MG, Peterson-Maduro J, Padberg Y, et al. Functional Dissection of the CCBE1 Protein: A Crucial Requirement for the Collagen Repeat Domain. Circ Res 2015;116:1660-9. [Crossref] [PubMed]
- Mesci A, Huang X, Taeb S, et al. Targeting of CCBE1 by miR-330-3p in human breast cancer promotes metastasis. Br J Cancer 2017;116:1350-7. [Crossref] [PubMed]
- Zhao YR, Liu H, Xiao LM, et al. The clinical significance of CCBE1 expression in human colorectal cancer. Cancer Manag Res 2018;10:6581-90. [Crossref] [PubMed]
- Mimeault M, Batra SK. Interplay of distinct growth factors during epithelial mesenchymal transition of cancer progenitor cells and molecular targeting as novel cancer therapies. Ann Oncol 2007;18:1605-19. [Crossref] [PubMed]
- Liu YM, Ge JY, Chen YF, et al. Combined Single-Cell and Spatial Transcriptomics Reveal the Metabolic Evolvement of Breast Cancer during Early Dissemination. Adv Sci (Weinh) 2023;10:e2205395. [Crossref] [PubMed]
- Biswas A, Ghaddar B, Riedlinger G, et al. Inference on spatial heterogeneity in tumor microenvironment using spatial transcriptomics data. Comput Syst Oncol 2022;2:e21043. [Crossref] [PubMed]
- Giannoni E, Taddei ML, Parri M, et al. EphA2-mediated mesenchymal-amoeboid transition induced by endothelial progenitor cells enhances metastatic spread due to cancer-associated fibroblasts. J Mol Med (Berl) 2013;91:103-15. [Crossref] [PubMed]
- Correa D, Somoza RA, Lin P, et al. Mesenchymal stem cells regulate melanoma cancer cells extravasation to bone and liver at their perivascular niche. Int J Cancer 2016;138:417-27. [Crossref] [PubMed]
- Sun Y, Xiao F, Sun H, et al. Transcriptome analysis of tumor-derived mesenchymal progenitor cells shows that CHST15 is a fibrosis regulator of retroperitoneal liposarcoma. Ann Transl Med 2022;10:360. [Crossref] [PubMed]
- He Y, Liu T, Dai S, et al. Tumor-Associated Extracellular Matrix: How to Be a Potential Aide to Anti-tumor Immunotherapy? Front Cell Dev Biol 2021;9:739161. [Crossref] [PubMed]
- Gonzalez-Velandia KY, Hernandez-Clavijo A, Menini A, et al. Expression pattern of Stomatin-domain proteins in the peripheral olfactory system. Sci Rep 2022;12:11447. [Crossref] [PubMed]
- Boimel PJ, Smirnova T, Zhou ZN, et al. Contribution of CXCL12 secretion to invasion of breast cancer cells. Breast Cancer Res 2012;14:R23. [Crossref] [PubMed]
- Ramos EA, Camargo AA, Braun K, et al. Simultaneous CXCL12 and ESR1 CpG island hypermethylation correlates with poor prognosis in sporadic breast cancer. BMC Cancer 2010;10:23. [Crossref] [PubMed]
- Wu W, Qian L, Chen X, et al. Prognostic significance of CXCL12, CXCR4, and CXCR7 in patients with breast cancer. Int J Clin Exp Pathol 2015;8:13217-24.
- Schelch K, Wagner C, Hager S, et al. FGF2 and EGF induce epithelial-mesenchymal transition in malignant pleural mesothelioma cells via a MAPKinase/MMP1 signal. Carcinogenesis 2018;39:534-45. [Crossref] [PubMed]
- Akl MR, Nagpal P, Ayoub NM, et al. Molecular and clinical significance of fibroblast growth factor 2 (FGF2 /bFGF) in malignancies of solid and hematological cancers for personalized therapies. Oncotarget 2016;7:44735-62. [Crossref] [PubMed]
- Teng Y, Guo B, Mu X, et al. KIF26B promotes cell proliferation and migration through the FGF2/ERK signaling pathway in breast cancer. Biomed Pharmacother 2018;108:766-73. [Crossref] [PubMed]
- Bos FL, Caunt M, Peterson-Maduro J, et al. CCBE1 is essential for mammalian lymphatic vascular development and enhances the lymphangiogenic effect of vascular endothelial growth factor-C in vivo. Circ Res 2011;109:486-91. [Crossref] [PubMed]
- Gordon-Weeks A, Lim SY, Yuzhalin A, et al. Tumour-Derived Laminin α5 (LAMA5) Promotes Colorectal Liver Metastasis Growth, Branching Angiogenesis and Notch Pathway Inhibition. Cancers (Basel) 2019;11:630. [Crossref] [PubMed]
- Zhang X, Li Q, Jiang W, et al. LAMA5 promotes human umbilical vein endothelial cells migration, proliferation, and angiogenesis and is decreased in preeclampsia. J Matern Fetal Neonatal Med 2020;33:1114-24. [Crossref] [PubMed]


