
Identification and validation of subclusters of papillary thyroid carcinoma based on Human Phenotype Ontology
Highlight box
Key findings
• We first identified subclusters in papillary thyroid carcinoma (PTC) based on the Human Phenotype Ontology (HPO) platform and found that patients with distinct subclusters have different prognoses. We then identified and validated the characteristic genes in the 4 subclusters.
What is known and what is new?
• Distinct subclusters of PTC are associated with different survival times and have different functional enrichment;
• Characteristic genes were identified in the 4 subclusters.
What is the implication, and what should change now?
• These findings are expected to serve as a crucial reference that will improve our understanding of PTC heterogeneity and the use of novel targets.
Introduction
Papillary thyroid carcinoma (PTC) is the most common subtype of thyroid cancer, which is the most common primary endocrine malignancy (1). The prognosis for PTC is generally excellent, with 10-year overall survival rates in the range of 80–95%; nevertheless, some patients will present advanced disease and will require targeted therapy (2,3). The increase in PTC diagnoses has prompted studies seeking to determine its pathogenesis, establish a diagnostic model of PTC, analyze the functional subclusters, and discover novel targets (4-6). Using multiple gene sets as sample characteristics for subtype clustering can improve the robustness of subtype clustering. For example, Hong et al. discovered four molecular subtypes of PTC, identified a 20-gene expression signature, which can predict the diagnosis of PTC (7). Li et al. found there was different somatic mutations and a unique transcriptomic signature in PTC based on multi-omics analysis (8). Another study explored whether alternative splicing events reflect new the molecular and histological subtypes of PTC, and found NUMA1_17515 and TUBB3_38175 were two alternative splicing biomarkers for PTC subclassification and characterization (9). The Human Phenotype Ontology (HPO) platform, which contains 5,142 gene sets, is available at http://www.gsea-msigdb.org/gsea/msigdb/human/genesets.jsp?collection=HPO. The HPO platform provides well-defined phenotypes in humans (10), and previous study has confirmed that it can predict genes from phenotypes (11). Additionally, HPO annotations can be performed for deep phenotyping to improve whole-exome sequencing evaluations in some rare diseases (12). However, the screening and validation of functional subclusters in PTC based on HPO have not been reported. We first used this platform to identify subclusters, and then performed an enrichment analysis to examine the key biological processes and pathways associated with the subclusters. A gene mutation analysis of the subclusters was also conducted. For each subcluster, the differentially expressed genes (DEGs) were selected and validated. Finally, a single-cell RNA-sequencing (scRNA-seq) data set was used to verify the DEGs. Our analysis demonstrated that distinct subclusters of PTC are associated with different survival times and have different functional enrichment. The DEGs identified in each subcluster are expected to serve as a crucial reference that will extend our understanding of PTC heterogeneity. We present this article in accordance with the STREGA reporting checklist (available at https://gs.amegroups.com/article/view/10.21037/gs-23-124/rc).
Methods
Patients
We used arrayExpress data containing the GSE3467, GSE3678, GSE6004, and GSE29265 data sets (which only included non-post-Chernobyl PTC data), which were downloaded from the Gene Expression Omnibus (GEO) database. GSE29265 comprising 10 tumor samples and 10 normal samples, GSE3467 comprising 9 tumor samples and 9 normal samples, GSE3678 comprising 7 tumor samples and 7 normal samples, GSE6004 comprising 14 tumor samples and 4 normal samples, were collected. Individual age and sex of the patients are listed in Table S1 and Table S2. Bulk RNA-sequencing (RNA-seq) data from the Genomic Data Commons-The Cancer Genome Atlas-Thyroid Carcinoma (GDC-TCGA-THCA) (https://xenabrowser.net/datapages/) were downloaded from University of Cingifornia Sisha Cruz (UCSC) Xena (13). Information of the normal samples in TCGA are listed in Table S3, information of tumor samples are listed in Table S4. The scRNA-seq data set GSE184362 was downloaded from the GEO database.
Data collection
The HPO terms were downloaded from C5 sub-collection HPO: Human Phenotype Ontology of the Molecular Signatures Database (MSigDB) (version 2022.1) (https://hpo.jax.org/app/) (10). In the R4.0.3 software, the “affy” R package (version 1.68.0) and “limma” package (version 3.46.0) were applied to integrate 4 arrayExpress data and for batch-effect correction (14,15). The annotation package “hgu133plus2.db” (version 3.2.3) was used to convert the probeset IDentities (IDs) into gene symbols (16). For the bulk RNA-seq data analysis, the read counts of the genes or transcripts were normalized using fragments per kilobase million, trans per million, and log2 transformation methods. The gencode.v22.annotation.gene.probeMap file was used for the genetic name conversion (13). The “Seurat” (version 4.3.0) and “harmony” (version 0.1.0) packages were used to integrate the multiple data and batch-effect corrections for the scRNA-seq data (17,18), and Uniform Manifold Approximation and Projection (UMAP) dimension reduction visualization was performed for the principal component analysis results based on the “harmony” package (version 0.1.0) (19).
Calculation of HPO scores in PTC
In R4.0.3 software, the PLAGE algorithm of the “GSVA” package (version 1.38.2) was used to calculate the HPO scores of the arrayExpress and bulk RNA-seq gene expression matrix, respectively (20-22). The Wilcoxon test was used to examine differences in the HPO scores obtained from the arrayExpress and bulk RNA-seq data between the PTC and normal samples. The screening threshold was a P value <0.0001. Finally, the intersection of the arrayExpress and bulk RNA-seq results was determined.
Clustering of subclusters in PTC using HPO
The package “ConsensusClusterPlus” (version 1.54.0) was used to generate subclusters based on the gene set scores at the intersection of the HPO gene sets (23). Then, k-means clustering was applied to analyze the subclusters of the PTC samples in the bulk RNA-seq data. The “Survival” (version 3.2.7) and “survminer” (version 0.4.9) packages were used to compare the prognosis between different subclusters (24-26).
Functional characterization of HPO subclusters in PTC
The “Limma” package (version 3.46.0) was used to analyze the genetic difference between the PTC subclusters and normal samples. The screening threshold was as follows: a P value after correction <0.05, and an absolute value of log-fold change >1. Subsequently, the “ClusterProfiler” package (version 3.18.1) was used to enrich and analyze the DEGs in the background of HPO, and the top 5 functions were visualized (25,27,28). Finally, the “maftools” package (version 2.6.5) was used in combination with the SNV analysis result file of THCA to visualize the mutation background of each subcluster (29).
Identification and validation of characteristic genes in the subclusters
The top 10 differentially downregulated and upregulated genes were selected to determine the intersection and create the Venn diagrams, and the unique and common genes among the 4 subclusters were selected for the subsequent analysis. Next, the “pheatmap” package (version 1.0.12) was used to display the correlations between the clusters and clinical parameters based on bulk RNA-seq data, which were validated using the arrayExpress data.
Verification of characteristic genes using the scRNA-seq data set
First, the “Seurat” package (version 4.3.0) was used to annotate the cell types from the scRNA-seq data (21), and the annotation reference was as follows: T cell markers (CD3D, CD3E, CD3G, and CD247), B cell markers [membrane spanning 4-domains A1 (MS4A1) and CD19], plasma cell markers [CD79A and X-box binding protein 1 (XBP1)], myeloid cell markers [integrin subunit alpha M (ITGAM), integrin subunit alpha X (ITGAX), and lysozyme (LYZ)], fibroblast markers [collagen type I alpha 1 chain (COL1A1), collagen type III alpha 1 chain (COL3A1), and actin alpha 2 (ACTA2)], endothelial cell markers [platelet and endothelial cell adhesion molecule 1 (PECAM1) and endoglin (ENG)], thyroid cell markers [keratin 18 (KRT18), keratin 8 (KRT8), and keratin 7 (KRT7)], and cell proliferation markers [marker of proliferation Ki-67 (MKI67) and DNA topoisomerase II alpha (TOP2A)]. Finally, the respective expression levels of these genes in the PTC cell types and normal samples were verified, and the “plot1cell” package was used for visualization.
Statistical analysis
All the analysis and the data visualization were performed using ggplot2 (version 3.4.0) in R4.0.3 software, except for those with special instructions used default parameters.
Overall survival prognosis was analyzed by Kaplan-Meier method, and difference was tested by log-rank t-test. In differential gene analysis, limma package lmFit was used for linear fitting of each gene to calculate gene difference multiples and standard errors, and then eBayes was used for empirical Bayesian smoothing of standard errors to obtain statistical values of differential test.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Identification of subclusters based on HPO
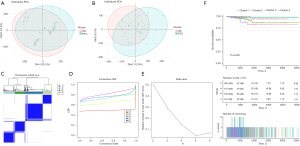
Firstly, we generated subclusters based on the gene set scores at the intersection of the HPO gene sets, via the package “ConsensusClusterPlus” (version 1.54.0). The results of the consistency cluster analysis showed that each cluster had a good differentiation degree when k =4 (Figure 1A-1E). Notably, when the patients were grouped into 4 subclusters, there was a statistically significant difference in the survival times of the patients. Specifically, patients in Clusters 1 and 3 had better survival times, while those in Cluster 4 had the worst survival time (Figure 1F). These results suggested that we had identified 4 subclusters based on HPO.
Functional characterization of the 4 subclusters
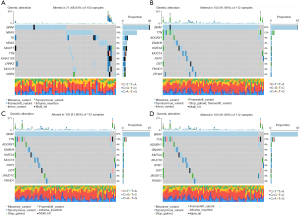
The downregulated genes in Cluster 1 were associated with functions such as brachydactyly, aplasia hypoplasia involving bones of the lower limbs, aplasia hypoplasia involving bones of the feet, an abnormality of the upper respiratory tract, and abnormal larynx morphology (Figure 2A). The upregulated genes in Cluster 1 were associated with functions such as proximal muscle weakness in the upper limbs, neurofibrillary tangles, the electromyography (EMG) decremental response of compound muscle action potential to repetitive nerve stimulation, dermatological manifestations of systemic disorders, and jaw muscle abnormality (Figure 2B). The downregulated genes in Cluster 2 were associated with functions such as thyroid defects in the oxidation and organification of iodide, hypoplasia of the epiglottis, elevated circulating thyroid-stimulating hormone concentration, abnormal thyroid-stimulating hormone levels, and abnormal circulating thyroglobulin levels (Figure 2C). The upregulated genes in Cluster 2 were associated with functions such as pustule, pulmonary fibrosis, palmar hyperhidrosis, abnormal pulmonary interstitial morphology, and abnormal pleura morphology (Figure 2D).

The downregulated genes in Cluster 3 were associated with functions such as thyroid defects in oxidation and organification of iodide, mastoiditis, hypokalemic alkalosis, calf musculature abnormality, and abnormal thyroid hormone levels (Figure 2E). The upregulated genes in Cluster 3 were associated with functions such as palmar hyperhidrosis, nail dystrophy, blistering by anatomical location, alopecia, and abnormal blistering of the skin (Figure 2F). Further, the enrichment analysis indicated that the downregulated genes in Cluster 4 were mainly involved in recurrent bacterial infections, immunodeficiency, decreased circulating immunoglobin G (IgG) levels, humoral immunity abnormality, and abnormal circulating IgG levels (Figure 2G). The upregulated genes in Cluster 4 were associated with functions such as status epilepticus without prominent motor symptoms, sparse body hair, non-convulsive status epilepticus without coma, bloody diarrhea, and abnormal urine calcium concentration (Figure 2H).
As Figure 3 shows, the clusters differed significantly in gene mutations, especially with respect to Cluster 1, which was completely different to the other 3 clusters. Cluster 1 contained 35% BRAF mutations, 18% NRAS mutations, 7% thyroglobulin (TG) mutations, 6% HRAS mutations, and 6% microtubule actin crosslinking factor 1 (MACF1) mutations. The top 5 mutated genes in the other 3 clusters were the BRAF, titin (TTN), adhesion G protein-coupled receptor V1 (ADGRV1), dynein axonemal heavy chain 9 (DNAH9), lysine methyltransferase 2A (KMT2A), and BRAF mutations accounted for 87%.
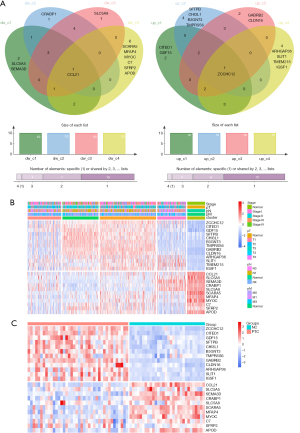
Identification of the characteristic genes in the 4 subclusters
The results showed that CCL21 was the most commonly downregulated gene in the 4 subclusters, and ZCCHC12 was the most commonly upregulated gene. We also found significant differences in the characteristic genes among the 4 clusters. In Cluster 1, the specific downregulated genes were solute carrier family 5 member 5 (SLC5A5) and semaphorin 3D (SEMA3D), while the specific upregulated genes were Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 (CITED1) and growth differentiation factor 15 (GDF15). In Cluster 2, the specific downregulated gene was cellular retinoic acid binding protein 1 (CRABP1), while the specific upregulated genes were surfactant protein B (SFTPB), chitinase 3 like 1 (CHI3L1), UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 (B3GNT3), and transmembrane serine protease 6 (TMPRSS6). In Cluster 3, the specific downregulated gene was solute carrier family 5 member 8 (SLC5A8), while the specific upregulated genes were gamma-aminobutyric acid type A receptor subunit beta2 (GABRB2) and claudin 16 (CLDN16). In Cluster 4, the specific downregulated genes were scavenger receptor class A member 5 (SCARA5), microfibril associated protein 4 (MFAP4), myocilin (MYOC), complement C7 (C7), secreted frizzled related protein 2 (SFRP2), and apolipoprotein D (APOD), while the specific upregulated genes were Rho GTPase activating protein 36 (ARHGAP36), slit guidance ligand 1 (SLIT1), transmembrane protein 215 (TMEM215), and immunoglobulin superfamily member 1 (IGSF1) (Figure 4A).
Validation of the characteristic genes in 4 subclusters
First, we verified the common or unique characteristic genes in the 4 subclusters based on the bulk RNA-seq data. The results showed that ZCCHC12, CITED1, GDF15, SFTPB, CHI3L1, B3GNT3, TMPRSS6, GABRB2, CLDN16, ARHGAP36, SLIT1, TMEM215, and IGSF1 expression levels were decreased in normal tissues and upregulated in PTC tissues. Further, CCL21, SLC5A5, SEMA3D, CRABP1, SLC5A8, SCARA5, MFAP4, MYOC, C7, SFRP2, and APOD expression levels were downregulated in PTC tissues compared to normal tissues (Figure 4B). Finally, we used the ArrayExpress data for verification, and found that these results were consistent with our aforementioned results, except that TMEM215 was not expressed in PTC tissues (Figure 4C).
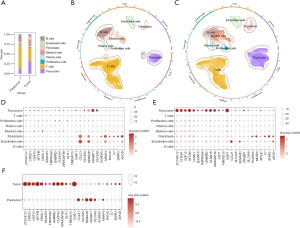
After the cell-type annotation of the scRNA-seq data, we found that the levels of thyrocytes, fibroblasts, and myeloid cells were increased in PTC, while those of T cells and B cells were decreased in PTC compared to the levels in normal tissues (Figure 5A-5C). Notably, we found that the characteristic genes in the 4 subclusters were mainly expressed in thyrocytes, endothelial cells, and fibroblasts, and were rarely expressed in immune cells (Figure 5D,5E). A further analysis showed that the downregulated gene CCL21 was mainly expressed in the endothelial cells and fibroblasts of the normal samples. In PTC, CCL21 was mainly expressed in the endothelial cells. Moreover, compared to that in the PTC tissues, there was a significant increase in CCL21 expression in the normal tissues. In addition, we found that ZCCHC12 was mainly expressed in the thyrocytes of PTC (Figure 5D-5F).
Discussion
Previous study has noted that HPO is widely available for the differential diagnosis of rare diseases, phenotype-driven investigations based on next-generation sequence-variation data, and translational studies (30). To the best of our knowledge, no previous study had examined the association between HPO and PTC. In this study, we identified subclusters based on HPO and found statistically significant differences in survival among the 4 subclusters, patients in Clusters 1 and 3 had better survival times, while those in Cluster 4 had the worst survival time (P=0.028). These results suggest that subclusters based on HPO can be used to predict the clinical prognosis of PTC.
The results of the enrichment analysis of these subclusters revealed that some of the subclusters were closely related to thyroid functions; for example, the downregulated genes in Cluster 3 were determined to be involved in thyroid defects in the oxidation and the organification of iodide, and abnormal thyroid hormone levels.
The present study also investigated whether CCL21 and ZCCHC12 were common down- or upregulated genes, respectively, in the 4 subclusters. The results supported evidence from previous observations, including those of Liu et al., who found that the expression of CCL21 is lower in PTC tissues than in Hashimoto’s thyroiditis tissues (31), and Smallridge et al., who reported that CCL21 is associated with lymphocyte infiltration and is differentially overexpressed in BRAF-wild-type tumors compared to BRAF V600E-mutation-harboring tumors (32).
A previous study revealed that ZCCHC12 expression is significantly upregulated in PTC, but no significant relationships were found between the expression of ZCCHC12 and the biochemical and clinicopathological features of PTC (33). Another important finding was that the expression of ZCCHC12 was upregulated and related to lymph node metastasis in primary PTC tumors, and PTC could be inhibited by downregulating ZCCHC12 (34). As patients in Clusters 1 and 3 had better survival, and SLC5A5, SEMA3D, and SLC5A8 expression levels were specifically downregulated in both clusters, the effect of these 3 genes in PTC is an important issue for future research.
Among the 22 characteristic genes in the 4 subclusters, the roles of SLC5A5, SEMA3D, CITED1, GDF15, CRABP1, SFTPB, CHI3L1, SLC5A8, GABRB2, SCARA5, SFRP2, and ARHGAP36 in PTC have been reported. For example, previous study has reported that SLC5A5 is expressed at a lower level in PTC, and the lower expression of SLC5A5 is correlated with aggressiveness and BRAF, NRAS, and TERTp mutations (35). The present study found that SLC5A5 was a specific downregulated gene in Cluster 1, which had a low percentage of BRAF mutations but a high percentage of NRAS mutations. Thus, the downregulation of SLC5A5 might be more closely related to NRAS mutations than to BRAF and TERTp mutations. Further, CITED1 and SFTPB are more highly expressed in PTC tissues than in follicular thyroid carcinoma and normal thyroid tissues (36), while the diagnostic utility of SFTPB and CITED1 was found to be poor in PTC (37). A recent study confirmed that CITED1 promotes PTC (38). SLC5A8 codes for a transporter belonging to the Na(+)/glucose co-transporter gene family, and the protein coded by SLC5A8 can transport iodide and regulate the Na(+)-coupled and electrogenic transport of many monocarboxylates (39). Our results support our earlier observations, which showed that SEMA3D (40), CRABP1 (41), SLC5A8 (42), and SCARA5 (43) are tumor suppressor genes in PTC, while GDF15 (44), CHI3L1 (45,46), GABRB2 (47), and ARHGAP36 (48) are oncogenic genes in PTC. However, very little research has been conducted on the roles of B3GNT3, TMPRSS6, CLDN16, MFAP4, MYOC, C7, APOD, SLIT1, TMEM215, and IGSF1 in PTC.
Notably, the characteristic genes in the 4 subclusters were mainly expressed in thyrocytes, endothelial cells, and fibroblasts. However, these genes were rarely expressed in immune cells. This finding suggests that patients with PTC would not benefit from immunotherapy.
Despite the promising results, a number of questions remain. For example, more experimental studies need to be conducted to establish predictive models based on HPO. Additionally, further research should be undertaken to investigate why the patients in Clusters 1 and 3 had better prognoses and to explore the unreported function of such characteristic genes in PTC.
Conclusions
We first used the HPO platform to identify the subclusters in PTC and demonstrated that patients with distinct subclusters of disease exhibit different prognoses, which enabled us to construct predictive models of patient prognosis. Additionally, we identified and validated the characteristic genes in the 4 subclusters that might play important roles in PTC. Our findings provide a crucial reference that will improve understandings of PTC heterogeneity.
Acknowledgments
Funding: This work was supported by the “10,000 Talents Plan” of Zhejiang Province (No. 2020R52029), the Natural Science Foundation of Zhejiang Province (Nos. LY22H160041, and LY22H160036), the National Natural Science Foundation of China (Nos. 82273287, 82204445, and 82203858), and the Zhejiang Medical Technology Plan Project (No. 2022KY060).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://gs.amegroups.com/article/view/10.21037/gs-23-124/rc
Peer Review File: Available at https://gs.amegroups.com/article/view/10.21037/gs-23-124/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://gs.amegroups.com/article/view/10.21037/gs-23-124/coif). JJD has received honoraria for lectures from Lilly, Faes, Menarini, MSD and Takeda; and received support for attending meetings and/or travel from Takeda, Menarini and Ipsen. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Filetti S, Durante C, Hartl D, et al. Thyroid cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann Oncol 2019;30:1856-83. [Crossref] [PubMed]
- Schlumberger M, Leboulleux S. Current practice in patients with differentiated thyroid cancer. Nat Rev Endocrinol 2021;17:176-88. [Crossref] [PubMed]
- Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014;159:676-90. [Crossref] [PubMed]
- Kanokwongnuwat W, Larbcharoensub N, Sriphrapradang C, et al. Risk-stratified papillary thyroid microcarcinoma: post-operative management and treatment outcome in a single center. Endocrine 2022;77:134-42. [Crossref] [PubMed]
- Ulisse S, Baldini E, Lauro A, et al. Papillary Thyroid Cancer Prognosis: An Evolving Field. Cancers (Basel) 2021;13:5567. [Crossref] [PubMed]
- Fallahi P, Ferrari SM, Galdiero MR, et al. Molecular targets of tyrosine kinase inhibitors in thyroid cancer. Semin Cancer Biol 2022;79:180-96. [Crossref] [PubMed]
- Hong S, Xie Y, Cheng Z, et al. Distinct molecular subtypes of papillary thyroid carcinoma and gene signature with diagnostic capability. Oncogene 2022;41:5121-32. [Crossref] [PubMed]
- Li Q, Feng T, Zhu T, et al. Multi-omics profiling of papillary thyroid microcarcinoma reveals different somatic mutations and a unique transcriptomic signature. J Transl Med 2023;21:206. [Crossref] [PubMed]
- Park J, Kim D, Lee JO, et al. Dissection of molecular and histological subtypes of papillary thyroid cancer using alternative splicing profiles. Exp Mol Med 2022;54:263-72. [Crossref] [PubMed]
- Köhler S, Gargano M, Matentzoglu N, et al. The Human Phenotype Ontology in 2021. Nucleic Acids Res 2021;49:D1207-17. [Crossref] [PubMed]
- Slavotinek A, Prasad H, Yip T, et al. Predicting genes from phenotypes using human phenotype ontology (HPO) terms. Hum Genet 2022;141:1749-60. [Crossref] [PubMed]
- Schön U, Holzer A, Laner A, et al. HPO-driven virtual gene panel: a new efficient approach in molecular autopsy of sudden unexplained death. BMC Med Genomics 2021;14:94. [Crossref] [PubMed]
- Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 2020;38:675-8. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Gautier L, Cope L, Bolstad BM, et al. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20:307-15. [Crossref] [PubMed]
- Zhan Z, Chen Y, Duan Y, et al. Identification of key genes, pathways and potential therapeutic agents for liver fibrosis using an integrated bioinformatics analysis. PeerJ 2019;7:e6645. [Crossref] [PubMed]
- Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573-87.e29. [Crossref] [PubMed]
- Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 2019;16:1289-96. [Crossref] [PubMed]
- Liu X, Xu X, Wu Z, et al. Integrated single-cell RNA-seq analysis identifies immune heterogeneity associated with KRAS/TP53 mutation status and tumor-sideness in colorectal cancers. Front Immunol 2022;13:961350. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Liang L, Yu J, Li J, et al. Integration of scRNA-Seq and Bulk RNA-Seq to Analyse the Heterogeneity of Ovarian Cancer Immune Cells and Establish a Molecular Risk Model. Front Oncol 2021;11:711020. [Crossref] [PubMed]
- Deng Y, Li K, Tan F, et al. Gene Model Related to m6A Predicts the Prognostic Effect of Immune Infiltration on Head and Neck Squamous Cell Carcinoma. J Oncol 2021;2021:1814266. [Crossref] [PubMed]
- Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26:1572-3. [Crossref] [PubMed]
- Zhu Z, Qin J, Dong C, et al. Identification of four gastric cancer subtypes based on genetic analysis of cholesterogenic and glycolytic pathways. Bioengineered 2021;12:4780-93. [Crossref] [PubMed]
- You W, Ouyang J, Cai Z, et al. Comprehensive Analyses of Immune Subtypes of Stomach Adenocarcinoma for mRNA Vaccination. Front Immunol 2022;13:827506. [Crossref] [PubMed]
- Zheng J, Zhang T, Guo W, et al. Integrative Analysis of Multi-Omics Identified the Prognostic Biomarkers in Acute Myelogenous Leukemia. Front Oncol 2020;10:591937. [Crossref] [PubMed]
- Wang L, Wang D, Yang L, et al. Cuproptosis related genes associated with Jab1 shapes tumor microenvironment and pharmacological profile in nasopharyngeal carcinoma. Front Immunol 2022;13:989286. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747-56. [Crossref] [PubMed]
- Groza T, Köhler S, Moldenhauer D, et al. The Human Phenotype Ontology: Semantic Unification of Common and Rare Disease. Am J Hum Genet 2015;97:111-24. [Crossref] [PubMed]
- Liu C, Pan Y, Li Q, et al. Bioinformatics analysis identified shared differentially expressed genes as potential biomarkers for Hashimoto's thyroiditis-related papillary thyroid cancer. Int J Med Sci 2021;18:3478-87. [Crossref] [PubMed]
- Smallridge RC, Chindris AM, Asmann YW, et al. RNA sequencing identifies multiple fusion transcripts, differentially expressed genes, and reduced expression of immune function genes in BRAF (V600E) mutant vs BRAF wild-type papillary thyroid carcinoma. J Clin Endocrinol Metab 2014;99:E338-47. [Crossref] [PubMed]
- Li QL, Chen FJ, Lai R, et al. ZCCHC12, a potential molecular marker of papillary thyroid carcinoma: a preliminary study. Med Oncol 2012;29:1409-17. [Crossref] [PubMed]
- Wang O, Zheng Z, Wang Q, et al. ZCCHC12, a novel oncogene in papillary thyroid cancer. J Cancer Res Clin Oncol 2017;143:1679-86. [Crossref] [PubMed]
- Tavares C, Coelho MJ, Eloy C, et al. NIS expression in thyroid tumors, relation with prognosis clinicopathological and molecular features. Endocr Connect 2018;7:78-90. [Crossref] [PubMed]
- Huang Y, Prasad M, Lemon WJ, et al. Gene expression in papillary thyroid carcinoma reveals highly consistent profiles. Proc Natl Acad Sci U S A 2001;98:15044-9. [Crossref] [PubMed]
- Nasr MR, Mukhopadhyay S, Zhang S, et al. Immunohistochemical markers in diagnosis of papillary thyroid carcinoma: Utility of HBME1 combined with CK19 immunostaining. Mod Pathol 2006;19:1631-7. [Crossref] [PubMed]
- Li H, Guan H, Guo Y, et al. CITED1 promotes proliferation of papillary thyroid cancer cells via the regulation of p21 and p27. Cell Biosci 2018;8:57. [Crossref] [PubMed]
- Ganapathy V, Gopal E, Miyauchi S, et al. Biological functions of SLC5A8, a candidate tumour suppressor. Biochem Soc Trans 2005;33:237-40. [Crossref] [PubMed]
- Hai R, You Q, Wu F, et al. Semaphorin 3D inhibits proliferation and migration of papillary thyroid carcinoma by regulating MAPK/ERK signaling pathway. Mol Biol Rep 2022;49:3793-802. [Crossref] [PubMed]
- Hawthorn L, Stein L, Varma R, et al. TIMP1 and SERPIN-A overexpression and TFF3 and CRABP1 underexpression as biomarkers for papillary thyroid carcinoma. Head Neck 2004;26:1069-83. [Crossref] [PubMed]
- Porra V, Ferraro-Peyret C, Durand C, et al. Silencing of the tumor suppressor gene SLC5A8 is associated with BRAF mutations in classical papillary thyroid carcinomas. J Clin Endocrinol Metab 2005;90:3028-35. [Crossref] [PubMed]
- Zheng C, Xia EJ, Quan RD, et al. Scavenger receptor class A, member 5 is associated with thyroid cancer cell lines progression via epithelial-mesenchymal transition. Cell Biochem Funct 2020;38:158-66. [Crossref] [PubMed]
- Kang YE, Kim JM, Lim MA, et al. Growth Differentiation Factor 15 is a Cancer Cell-Induced Mitokine That Primes Thyroid Cancer Cells for Invasiveness. Thyroid 2021;31:772-86. [Crossref] [PubMed]
- Cheng SP, Lee JJ, Chang YC, et al. Overexpression of chitinase-3-like protein 1 is associated with structural recurrence in patients with differentiated thyroid cancer. J Pathol 2020;252:114-24. [Crossref] [PubMed]
- Luo D, Chen H, Lu P, et al. CHI3L1 overexpression is associated with metastasis and is an indicator of poor prognosis in papillary thyroid carcinoma. Cancer Biomark 2017;18:273-84. [Crossref] [PubMed]
- Jin Y, Jin W, Zheng Z, et al. GABRB2 plays an important role in the lymph node metastasis of papillary thyroid cancer. Biochem Biophys Res Commun 2017;492:323-30. [Crossref] [PubMed]
- Yan T, Qiu W, Song J, et al. ARHGAP36 regulates proliferation and migration in papillary thyroid carcinoma cells. J Mol Endocrinol 2021;66:1-10. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


