
The ctDNA-based postoperative molecular residual disease status in different subtypes of early-stage breast cancer
Highlight box
Key findings
• We divided patients into 4 molecular subtypes based on the expression of hormone receptor (HR) and hormone epidermal receptor 2 (HER2) status. The proportion of molecular residual disease (MRD) detected in different molecular subtypes of early-stage breast cancer is inequable.
•
What is known and what is new?
• Somatic mutations in tissue of patients with early-stage breast cancer were well studied.
• We revealed the status of MRD in different molecular types of breast cancer in China. This is the first study to analyse the difference of somatic mutations in tissues based on MRD status.
What is the implication, and what should change now?
• Genetic mutations in tissues may be biomarkers of MRD status. Large-scale studies with survival analysis are needed to further explore our findings.
Introduction
Breast cancer is one of the leading causes of cancer-related deaths in women worldwide with an estimated 0.68 million deaths in 2020 (1). Previous study has confirmed that breast cancer is a highly heterogeneous disease (2). Different molecular subtypes of breast cancer have different biological behaviors, which directly affect the clinical treatment decision-making and prognosis evaluation (3). Currently, the most commonly used molecular subtype is defined by the absence or over expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) according to immunohistochemistry (IHC) or fluorescence in situ hybridization (FISH). Reports have shown that 70–80% of early-stage, non-metastatic breast cancers are considered curable (3). Therefore, the early diagnosis of breast cancer and the early detection of tumor recurrence and metastasis are of great significance. More adequate biomarkers to identify individuals at high risk of recurrence are needed.
Circulating tumor DNA (ctDNA) isolated from plasma contains genetic mutations that can be representative of the whole picture of tumor-derived alterations (4). The National Comprehensive Cancer Network (NCCN) guidelines have introduced ctDNA as new marker for risk of recurrence in early-stage colon cancer. The detection of ctDNA in postoperative patients is considered molecular residual disease (MRD), which reflects the presence of residual tumor cells or micro-metastases in patients who lack radiological signs after radical dissection (5). Several approaches have been reported for detection of MRD in solid tumors, 2 of which are commonly used: one is based on polymerase chain reaction (PCR), and the other is next-generation sequencing (NGS). From another perspective, based on whether the primary tumor-derived alterations need to be tested, MRD has two major technical routes, namely, tumor-informed assays and tumor-agnostic assays. Regardless of the technology used for the analysis of MRD, a detection of ctDNA in peripheral blood obtained after surgery or adjuvant therapy, or during follow-up represents high risk of recurrence (6).
In 2016, a study showed that among 55 early breast cancer patients, the disease-free survival (DFS) of MRD-positive (ctDNA detected) patients was shorter than that of MRD-negative (ctDNA not detected) patients (6.5 months vs. median not reached) (7). Multiple studies have shown that using ctDNA to identify MRD can predict patients’ recurrence risk with high sensitivity in diverse cancers such as breast cancer, pancreatic cancer and colorectal cancer (7-9). The lead time from ctDNA-based detection of MRD to clinical relapse will enable individual follow-up and intervention therapy for curative treatment (9). However, there are few studies on postoperative MRD as a prognostic and predictive biomarker in breast cancer, especially in the Chinese breast cancer population. In this study, we retrospectively analyzed the mutations of primary tumor tissues and peripheral blood in 82 cases of breast cancer. We report the findings of a real-world study to reveal the MRD of breast cancer patients with different molecular types in China. We present the following article in accordance with the MDAR reporting checklist (available at https://gs.amegroups.com/article/view/10.21037/gs-22-634/rc).
Methods
Patients and samples
In this retrospective cohort study, a cohort of 82 patients with stage I–III breast cancer was enrolled at The First Affiliated Hospital of Soochow University from December 2018 to July 2022. All patients received standard care and management, including surgery, adjuvant therapy, and follow up. The study was approved by the Ethics Committee of The First Affiliated Hospital of Soochow University (No. 2022013), and all cases provided written informed consent before undergoing any study-related procedures. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Surgically resected tissues were the main source of tumor tissue sampling for the DNA sequencing, and tissue obtained by puncture were also accessible for a portion of patients. Peripheral blood samples (20 mL) within 1 month after surgery and before initiation of adjuvant treatment were collected from the majority of cases; 2 patients had blood collected after adjuvant therapy. All peripheral blood samples were collected in Streck tubes (Streck, Omaha, NE, USA).
Sample processing and DNA extraction
Peripheral blood was processed within 3 days to separate plasma and white blood cells (WBCs). Pathological assessment was performed on all tissue specimens to evaluate sample quality, which required at least 20% tumor cell content. Circulating cell-free DNA (cfDNA) was isolated from the plasma using a QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany). Genomic DNA (gDNA) from WBCs and tumor tissues were extracted using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). All procedures for sample extraction were performed according to previous studies (10,11).
DNA sequencing and data analysis
The Qubit fluorometer (Invitrogen, Carlsbad, CA, USA) and the Qubit dsDNA HS (High Sensitivity) Assay Kit (Invitrogen) were used to quantify concentration of gDNA and cfDNA. The genome DNA was cut into 200–250 bp fragments by Covaris S2 instrument (Woburn, MA, USA). Subsequently, both genome DNA and cfDNA were assessed and constructed sequencing libraries. As reported in our previous study (12), sequencing libraries were constructed using the TruSeq DNA kits according to the manufacturer’s instruction. All DNA libraries from cases were detected by NGS based on a pan-cancer 1,021-gene panel, which were performed on the Illumina Nextseq CN 500 (Illumina, San Diego, CA, USA) or the Gene + Seq-2000 Sequencing System (GenePlus-Suzhou, Suzhou, China), according to the manufacturer instructions (11).
Sequencing data analysis
The clean data reads were mapped to the reference human genome (GRCh37) using the default parameters in Burrows-Wheeler aligner after removing terminal adaptor and low-quality reads as described in previous study (13). MuTect2 and Genome Analysis Toolkit (GATK) were employed to identify single nucleotide variants (SNVs) and small insertions and deletions (Indels), separately (12). Copy number alterations were identified with Contra algorithm (Version 2.0.8). ExAc and 1000 Genomes Project and Cosmic were used in mutation identification and filtering. The default parameters were used for all software or tools for sequencing data analysis. The ctDNA positivity was defined when at least one mutation had been detected in the plasma sample.
Statistical analyses
Fisher’s exact tests were used to analyze postoperative ctDNA positivity and molecular subtype. Most of the statistical analyses were performed using GraphPad Prism 8.0.2 (GraphPad Software, Inc., San Diego, CA, USA) in the study. The SNV count was defined as the total number of SNV events per sample. The Fisher’s t-test was performed to compare differences between various groups (14).
Results
Patient demographics
In total, 82 breast cancer cases, comprising 19 stage I, 44 stage II, and 19 stage III, were enrolled in this study. Some 13 patients received neoadjuvant therapy, and the remaining 69 patients were resectable. All patients were pathologically diagnosed with primary breast cancer, predominantly T2 (approximately 63%), with a median tumor size of 2.5 cm (range, 0.7–7 cm). In the study group, all patients were female, with a median age of 45 years (range, 29–79 years). A total of 82 tumor tissues and 224 plasma samples were collected in this real-world study. All patients had undergone surgical treatment. The clinicopathological characteristics are summarized in Table 1.
Table 1
| Characteristic | Patients (n=82) |
|---|---|
| Age, median [range] years | 45 [29–79] |
| Subtype | |
| HR+/HER2− | 37 (45%) |
| HR+/HER2+ | 16 (20%) |
| HR−/HER2− | 16 (20%) |
| HR−/HER2+ | 13 (15%) |
| Tumor size, median [range] cm | 2.5 [0.7–7] |
| Stage | |
| I | 19 (23%) |
| II | 44 (54%) |
| III | 19 (23%) |
| N stage | |
| N0 | 45 (55%) |
| N1–N3 | 37 (45%) |
| Family history | 21 (26%) |
HR, hormone receptor; HER2, human epidermal growth factor receptor 2.
The detection of MRD at different subtypes
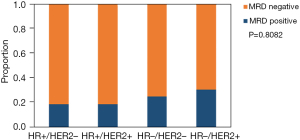
The 82 participants were classified into 4 subgroups according to the expression of ER, PR, or HER2. The expressions included HR+/HER2−, HR+/HER2+, HR−/HER2+, and HR−/HER2−, respectively. Postoperative blood samples were collected after surgery, and most of the patients preserved blood samples before adjuvant therapy. MRD positivity was defined as the detection of ctDNA at any time point after surgery. Among all cases, MRD positivity was detected in 18 of 82 patients (22.0%), and among the 4 subgroups, 18.9% were HR+/HER2−, 18.8% were HR+/HER2+, 25.0% were HR−/HER2−, and 30.8% were HR−/HER2+, respectively. The positive detection of MRD was more likely to be presented in the HR−/HER2+ subgroup (P=0.8082; as shown in Figure 1). Subsequent analyses were based on the MRD status after surgery.
Mutational profiling of tumor tissue specimens
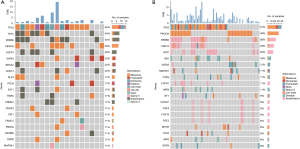
All samples including tumor, plasma, and matched blood cells were detected by a target 1,021-gene panel sequencing. A total of 849 somatic alterations were identified in 82 tissue samples, with a median of 10 mutations per patient (range, 1–41 mutations). The 849 somatic mutations consisted of 535 SNVs or indels, 302 copy number variants (CNVs), and 12 structural variations (SVs). Cases were divided in two groups according to these MRD statuses. The detected mutations in tumor tissue of the two groups are shown in Figure 2. In both the MRD negative and MRD positive subgroup, TP53 was the most frequently mutated gene, with a mutation frequency of 61% and 83%, respectively. Besides, we found that MYC amplification (44%), ERBB2 amplification (44%), and PIK3CA (39%) were the most frequently mutated genes (Figure 2A) in the MRD positive subgroup. Meanwhile, variations in PIK3CA (48%) and ERBB2 (36%) were common high-frequency mutated genes in the MRD-negative group (Figure 2B).
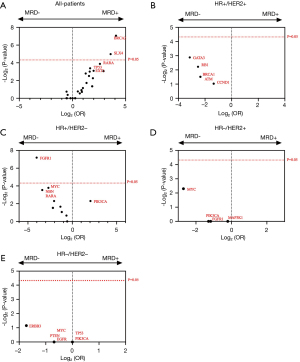
We attempted to analyze whether there were genes in tumor tissue that associated with the positive status of MRD. As shown in Figure 3, BRCA2 and SLX4 were significantly mutated in MRD-positive compared to MRD-negative groups. Patients with BRCA2 or SLX4 gene mutations in tissue samples were more likely to present an MRD positive status after surgery. Subsequently, the same comparison was performed in 4 different molecular subgroups. A total of 141, 170, 105, and 73 genes in the HR+/HER2−, HR+/HER2−, HR−/HER2+, and HR−/HER2− groups were included in this analysis, respectively. The FGFR1 gene differed significantly between the MRD positive and negative groups in the HR+HER2− group (Figure 3C). No significant difference was obtained in the other 3 subgroups, including HR+/HER2+, HR−/HER2+, and HR−/HER2−.
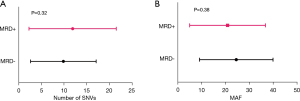
We further explored whether there were differences in the mutation load and max allele frequency (MAF) of mutations in the 2 MRD status groups. The SNV number count in the tissue sample of MRD-positive patients was higher than that of MRD-negative patients (mean 11.94 vs. 9.91 SNVs/sample, P=0.3293). The mean MAF in MRD-positive cases was lower than that of the MRD-negative group (20.90% vs. 24.54%, P=0.3809) (Figure 4).
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis
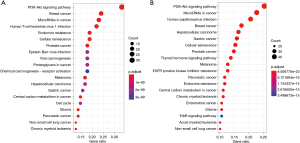
In order to understand the functional mechanism of the detected gene mutations in breast cancer with MRD-positive or MRD-negative, KEGG pathway enrichment analysis was performed based on the Metascape database as described previously (15). All somatic mutations involved in the 125 and 259 gene from 18 MRD-positive patients or 64-negative patients were included in KEGG enrichment analysis, respectively (Figure 5). Gene enrichment analysis showed that showed that the MRD-positive subgroup was mainly enriched in the “PI3K-Akt signaling pathway”, “Breast cancer”, “MicroRNAs in cancer”, “Human T-cell leukemia virus 1 infection”, “Endocrine resistance”, and “Cellular senescence”, yet the MRD-positive subgroup was mainly enriched in the “PI3K-Akt signaling pathway”, “MicroRNAs in cancer”, “Human papillomavirus infection”, “Breast cancer”, “Hepatocellular carcinoma”, and “Gastric cancer”. The above results highlight that the pathways for KEGG enrichment in MRD positive group are similar to those in the MRD-negative group, which indicate that there is no strong correlation between MRD status and the biological function of the mutated gene in the tissue.
Discussion
It is now well known that ctDNA is a component of cell-free DNA that is shed by tumors into the peripheral blood and the detectable MRD based on ctDNA after radical treatment is indicative of the presence of tumor in the body. Nevertheless, the previous data had demonstrated that the MAF in early stage was lower than that in advanced tumors (16). In the context of MRD, detection of ctDNA in the peripheral blood or other bodily fluids from tumor patients is more challenging. But it is technically feasible by technical methods such as NGS or digital droplet PCR (ddPCR) assays. Among the MRD detection assays, NGS is widely used on clinical practice because it can detect more mutations once. Meanwhile, there are several methods to increase sensitivity of mutation detection by NGS. As reported, tracking more mutations (17), higher DNA input (more peripheral blood volume) and increased number of MRD tests, increase sequencing depth (18,19) might be helpful for detection of low-frequency variants of ctDNA which means that the sensitivity of the ctDNA detection by NGS assay is increased. In this study, paired tissue samples and plasma samples were detected by the NGS panel containing 1,021 cancer-associated genes with a sequencing depth over 500× and 10,000×, respectively. We balanced various factors such as sequencing depth and mutation coverage to identify low-frequency variants to detect the presence MRD.
Breast cancer have caused the majority of cancer-related death worldwide (20,21). Adjuvant therapy for early breast cancer is mainly based on radical surgery combined with systemic therapy. Previously, decision-making of adjuvant therapy mostly relied on tumor stage, risk of recurrence and metastasis, and molecular type of the tumor (22). Multigene profiling assays on tissue and postoperative blood during follow-up play an important role in assisting clinical decision-making, which can help to balance the efficacy and disadvantages of adjuvant therapy, avoid over- or inadequate treatment, and thus benefit patients’ survival and quality of life. Several high-quality studies have revealed that MRD status based on ctDNA analysis is associated with patients’ prognostic outcomes for both relapse-free survival and overall survival in pan-cancers (e.g., lung cancer, colon cancer, breast cancer) in recent years (11,23,24). On the one hand, ctDNA is highly tumor specific and can accurately detect the presence of molecular residual disease almost in any solid tumors. What’s more, there is a lead time from ctDNA-positive detection to radiological recurrence. A study declared the median lead time between ctDNA detection and relapse was 10.7 months in 101 breast cancer (9), which suggesting that ctDNA could be a great adjunct to CT imaging for disease monitor. On the other hand, MRD has excellent prognostic value in breast cancer. Of the 101 female breast cancer, median relapse free survival (RFS) in patients with MRD detectable was shorter than that with MRD undetectable (median not reached) [hazard ratio (HR) =16.7; P<0.001] (9,25). CtDNA detection in 142 patients who had been diagnosed as stage 0-III breast cancer at the post-operative (HR =5.1) had inferior RFS (HR =5.1; P=0.00048) (18). As a result, the previous studies have focused on the association between MRD status and prognosis, and most of these studies and patient enrollment have been prospectively designed (26,27). However, few studies have combined MRD with HR/HER2 status to define different subtypes of breast cancer, and explored the genetic characteristics among the subgroups in tumor tissue. In the present study, we conducted a real-world retrospective study; all patients and samples were reexamined and collected according to both clinical follow-up and patients’ own wishes, which might better reflect the real situation of breast cancer in China.
Currently, breast cancer is classified into different molecular subtypes, which have great significance for individualized intervention and prognostic prediction (2,28,29). Different molecular subtypes were also associated with anti-tumor treatment resistance in advanced cancers (30-32). In our study, we divided patients into 4 molecular subtypes based on the expression of HR and HER2 status as described above. The four different subtypes of breast cancer had inequable clinicopathological characteristics and prognosis. As reported, patients with HR+ tend to have longer survival than that with HR- disease. HR-/Her2- is associated with worse survival. Compared to patients with HR+/HER2− disease, patients with ER+/HER2+ disease had a lower odds to develop radiographic ascites (33,34). The sensitivity to endocrine therapy was higher in HR+ patients than in HR− patients, especially in the HR+/HER2− group (35). Meanwhile, we classified all of the patients as either MRD-negative or MRD-positive based on the presence of ctDNA at postoperative surveillance. Analysis of tumor-derived mutations had enormous potential applications in cancer management, including earlier diagnosis of cancer, identification of driver alterations, monitoring of treatment response and detection of resistance mechanisms (36-38). TP53 mutations are common in breast cancer. As reported, TP53 mutations are detected in approximately 53.6% of breast cancer cases, and patients with TP53 mutations have a shorter DFS than those without TP53 mutations (28.5 vs. 40.6 months, P=0.005) (39). In this study, 65% of cases harbored TP53 gene alteration and the MRD-positive group had higher TP53 mutations than the MRD-negative group (83% vs. 61%, P=0.087), which is consistent with the worse prognosis among MRD-positive patients in the previous study. In addition, the MRD-positive group had a higher frequency of MYC amplification than the MRD-negative group (44% vs. 25%, P=0.109), which is also associated with high recurrence risk or metastasis in breast cancer (40).
Significant difference analysis in somatic mutations was performed on all patients and on the 4 HR/HER2 defined molecular subtypes, respectively. BRCA2 and SLX4 were found to be significantly associated with MRD-positive status among all the patients, with the detection rates of 44% and 17%, respectively. Interestingly, among the tissue samples of the 64 MRD-negative patients, only one patient harbored BRCA2 or SLX4 mutations. It is well known that BRCA2 is an important tumor suppressor gene that inhibits the proliferation, migration, and invasion of breast tumor cells. Zhong et al. summarized the association between BRCA mutations and survival in breast cancer and revealed that BRCA2 was not associated with breast cancer prognosis (41). Nevertheless, another study reported that the DFS of the BRCA1/2 mutant group was significantly better than that of the BRCA1/2 wild group in triple-negative breast cancer (42). The 5-year risk for contralateral breast cancer is higher in patients with BRCA2 mutations than that in non-BRCA carriers (9% vs. 3%, P<0.001) (43). Meanwhile, SLX4, a protein involved in DNA damage repair, is well studied in the FANCP subtype of Fanconi anemia patients with germline mutation. It has been reported that mutations on DNA damage response (DDR) genes are significantly associated with higher tumor mutation burden and worse prognosis in metastatic breast cancer (44). Numerous mutations in the SLX4 gene have been reported to be likely oncogenic, such as SLX4 p.P28Lfs*16 and breast invasive ductal carcinoma (cBioPortal database). In our group of MRD-positive patients, SLX4 mutations were detected in 4 of 18 patients. These mutations have not been clearly reported, and more studies may need to confirm the association between the SLX4 gene alternation and the pathogenic mechanism of breast cancer. In brief, both BRCA2 and SLX4 are involved in DNA damage repair, which may result in poor prognosis in MRD-positive patients harboring BRCA or SLX4 mutations. These inferences should be further explored and validated in larger-scale studies.
Among the other 4 molecular subgroups, significantly different genes were found only in the HR+/HER2− group. FGFR1 amplification was significantly enriched in the MRD-negative group in HR+/HER2−. FGFR genetic aberration was associated with poor prognosis and could increase the risk of brain metastases in breast cancer (45,46). Another report also mentioned that the overamplification/activation of FGFR1-4 could drive oncogenesis in cancers (47). Amina et al. proposed that FGFR1 expression was associated with distant metastasis in luminal B tumors but not with luminal A (HR+/HER2−) tumors (P=0.035). Our results also infer that patients with tissues with FGFR1 rearrangement-positive are more likely to present with a negative postoperative MRD and thus have a low risk of recurrence (48). Unfortunately, we did not obtain the survival of the patients at this stage and could not conduct more association analysis between gene mutations and survival. We will perform survival analysis in future studies.
There are some limitations to this study. Firstly, the size of the study population was too small to perform several analyses of the four subgroups. The statistical non-significance in our results of the analysis were also attributed to the small study population (49). Secondly, survival data are absent in the study, so we could not explore the relationship between MRD status or molecular subtype or certain gene and prognosis.
Conclusions
In this real-world study, we analyzed the MRD status in early-stage breast cancer. The proportion of MRD detected by different molecular subtypes of breast cancer is inequable. BRCA2 and SLX4 gene alteration may be associated with MRD positivity. Large-scale studies with survival analysis are needed to further explore our findings.
Acknowledgments
Funding: The present study was supported by the 24th Soochow University Extra-curricular academic research fund (No. KY20220121A) and the Guangzhou Municipal Science and Technology Planning Project (No. 202102020049).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://gs.amegroups.com/article/view/10.21037/gs-22-634/rc
Data Sharing Statement: Available at https://gs.amegroups.com/article/view/10.21037/gs-22-634/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://gs.amegroups.com/article/view/10.21037/gs-22-634/coif). All authors report funding support from the 24th Soochow University Extra-curricular academic research fund (No. KY20220121A) and the Guangzhou Municipal Science and Technology Planning Project (No. 202102020049). YLS, YGH and DJG report personal fees from Geneplus-Beijing. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics Committee of The First Affiliated Hospital of Soochow University (No. 2022013), and all cases provided written informed consent before undergoing any study-related procedures. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Yi Z, Ma F, Li C, et al. Landscape of somatic mutations in different subtypes of advanced breast cancer with circulating tumor DNA analysis. Sci Rep 2017;7:5995. [Crossref] [PubMed]
- Harbeck N, Penault-Llorca F, Cortes J, et al. Breast cancer. Nat Rev Dis Primers 2019;5:66. [Crossref] [PubMed]
- Wang Y, Zhao C, Chang L, et al. Circulating tumor DNA analyses predict progressive disease and indicate trastuzumab-resistant mechanism in advanced gastric cancer. EBioMedicine 2019;43:261-9. [Crossref] [PubMed]
- Pantel K, Alix-Panabières C. Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat Rev Clin Oncol 2019;16:409-24. [Crossref] [PubMed]
- Yang J, Gong Y, Lam VK, et al. Deep sequencing of circulating tumor DNA detects molecular residual disease and predicts recurrence in gastric cancer. Cell Death Dis 2020;11:346. [Crossref] [PubMed]
- Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015;7:302ra133. [Crossref] [PubMed]
- Jiang J, Ye S, Xu Y, et al. Circulating Tumor DNA as a Potential Marker to Detect Minimal Residual Disease and Predict Recurrence in Pancreatic Cancer. Front Oncol 2020;10:1220. [Crossref] [PubMed]
- Reinert T, Henriksen TV, Christensen E, et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol 2019;5:1124-31. [Crossref] [PubMed]
- Shen F, Liang N, Fan Z, et al. Genomic Alterations Identification and Resistance Mechanisms Exploration of NSCLC With Central Nervous System Metastases Using Liquid Biopsy of Cerebrospinal Fluid: A Real-World Study. Front Oncol 2022;12:889591. [Crossref] [PubMed]
- Zhang JT, Liu SY, Gao W, et al. Longitudinal Undetectable Molecular Residual Disease Defines Potentially Cured Population in Localized Non-Small Cell Lung Cancer. Cancer Discov 2022;12:1690-701. [Crossref] [PubMed]
- Lan J, Zhou YH, Zhang MX, et al. Molecular profiles and circulating tumor-DNA detected in Chinese early stage breast cancer. Gland Surg 2022;11:319-29. [Crossref] [PubMed]
- Peng W, Li B, Li J, et al. Clinical and genomic features of Chinese lung cancer patients with germline mutations. Nat Commun 2022;13:1268. [Crossref] [PubMed]
- Deng Z, Cui L, Li P, et al. Genomic comparison between cerebrospinal fluid and primary tumor revealed the genetic events associated with brain metastasis in lung adenocarcinoma. Cell Death Dis 2021;12:935. [Crossref] [PubMed]
- Duan Y, Du Y, Gu Z, et al. Expression, Prognostic Value, and Functional Mechanism of the KDM5 Family in Pancreatic Cancer. Front Cell Dev Biol 2022;10:887385. [Crossref] [PubMed]
- Chin RI, Chen K, Usmani A, et al. Detection of Solid Tumor Molecular Residual Disease (MRD) Using Circulating Tumor DNA (ctDNA). Mol Diagn Ther 2019;23:311-31. [Crossref] [PubMed]
- Parsons HA, Rhoades J, Reed SC, et al. Sensitive Detection of Minimal Residual Disease in Patients Treated for Early-Stage Breast Cancer. Clin Cancer Res 2020;26:2556-64. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Swanton C. Early stage NSCLC - challenges to implementing ctDNA-based screening and MRD detection. Nat Rev Clin Oncol 2018;15:577-86. [Crossref] [PubMed]
- Ye LF, Huang ZY, Chen XX, et al. Monitoring tumour resistance to the BRAF inhibitor combination regimen in colorectal cancer patients via circulating tumour DNA. Drug Resist Updat 2022;65:100883. [Crossref] [PubMed]
- Narayanan S, Cai CY, Assaraf YG, et al. Targeting the ubiquitin-proteasome pathway to overcome anti-cancer drug resistance. Drug Resist Updat 2020;48:100663. [Crossref] [PubMed]
- Liu P, Wang Z, Ou X, et al. The FUS/circEZH2/KLF5/ feedback loop contributes to CXCR4-induced liver metastasis of breast cancer by enhancing epithelial-mesenchymal transition. Mol Cancer 2022;21:198. [Crossref] [PubMed]
- Harris LN, Ismaila N, McShane LM, et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women With Early-Stage Invasive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol 2016;34:1134-50. [Crossref] [PubMed]
- Tie J, Cohen JD, Lahouel K, et al. Circulating Tumor DNA Analysis Guiding Adjuvant Therapy in Stage II Colon Cancer. N Engl J Med 2022;386:2261-72. [Crossref] [PubMed]
- Cai Z, Chen G, Zeng Y, et al. Comprehensive Liquid Profiling of Circulating Tumor DNA and Protein Biomarkers in Long-Term Follow-Up Patients with Hepatocellular Carcinoma. Clin Cancer Res 2019;25:5284-94. [Crossref] [PubMed]
- Erin N, Grahovac J, Brozovic A, et al. Tumor microenvironment and epithelial mesenchymal transition as targets to overcome tumor multidrug resistance. Drug Resist Updat 2020;53:100715. [Crossref] [PubMed]
- Garcia-Murillas I, Chopra N, Comino-Méndez I, et al. Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol 2019;5:1473-8. [Crossref] [PubMed]
- Zhou Y, Xu Y, Wang C, et al. Serial circulating tumor DNA identification associated with the efficacy and prognosis of neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat 2021;188:661-73. [Crossref] [PubMed]
- Yu Z, Lan J, Li W, et al. Circular RNA hsa_circ_0002360 Promotes Proliferation and Invasion and Inhibits Oxidative Stress in Gastric Cancer by Sponging miR-629-3p and Regulating the PDLIM4 Expression. Oxid Med Cell Longev 2022;2022:2775433. [Crossref] [PubMed]
- Nussinov R, Tsai CJ, Jang H. Anticancer drug resistance: An update and perspective. Drug Resist Updat 2021;59:100796. [Crossref] [PubMed]
- Chen D, Wang R, Yu C, et al. FOX-A1 contributes to acquisition of chemoresistance in human lung adenocarcinoma via transactivation of SOX5. EBioMedicine 2019;44:150-61. [Crossref] [PubMed]
- Zhu YQ, Zhou NH, Xu YW, et al. Genome-wide analysis of Chinese keloid patients identifies novel causative genes. Ann Transl Med 2022;10:883. [Crossref] [PubMed]
- Jiang M, Qi F, Zhang K, et al. MARCKSL1-2 reverses docetaxel-resistance of lung adenocarcinoma cells by recruiting SUZ12 to suppress HDAC1 and elevate miR-200b. Mol Cancer 2022;21:150. [Crossref] [PubMed]
- Huppert LA, Walker Z, Li M, et al. Clinical characteristics and outcomes in patients with metastatic breast cancer and pseudocirrhosis: a single center retrospective cohort study. Breast Cancer Res Treat 2022; Epub ahead of print. [Crossref]
- Wang X, Jiang W, Du Y, et al. Targeting feedback activation of signaling transduction pathways to overcome drug resistance in cancer. Drug Resist Updat 2022;65:100884. [Crossref] [PubMed]
- Loibl S, Poortmans P, Morrow M, Denkert C, Curigliano G. Breast cancer. Lancet 2021;397:1750-69. Erratum in: Lancet 2021;397:1710. [Crossref] [PubMed]
- Rizzo A, Ricci AD, Tavolari S, et al. Circulating Tumor DNA in Biliary Tract Cancer: Current Evidence and Future Perspectives. Cancer Genomics Proteomics 2020;17:441-52. [Crossref] [PubMed]
- Xavier CPR, Belisario DC, Rebelo R, et al. The role of extracellular vesicles in the transfer of drug resistance competences to cancer cells. Drug Resist Updat 2022;62:100833. [Crossref] [PubMed]
- Chen J, Zhang K, Zhi Y, et al. Tumor-derived exosomal miR-19b-3p facilitates M2 macrophage polarization and exosomal LINC00273 secretion to promote lung adenocarcinoma metastasis via Hippo pathway. Clin Transl Med 2021;11:e478. [Crossref] [PubMed]
- Yi Z, Ma F, Rong G, et al. Clinical spectrum and prognostic value of TP53 mutations in circulating tumor DNA from breast cancer patients in China. Cancer Commun (Lond) 2020;40:260-9. [Crossref] [PubMed]
- Stradella A, Gargallo P, Cejuela M, et al. Genomic characterization and tumor evolution in paired samples of metaplastic breast carcinoma. Mod Pathol 2022;35:1066-74. [Crossref] [PubMed]
- Zhong Q, Peng HL, Zhao X, et al. Effects of BRCA1- and BRCA2-related mutations on ovarian and breast cancer survival: a meta-analysis. Clin Cancer Res 2015;21:211-20. [Crossref] [PubMed]
- Fasching PA, Loibl S, Hu C, et al. BRCA1/2 Mutations and Bevacizumab in the Neoadjuvant Treatment of Breast Cancer: Response and Prognosis Results in Patients With Triple-Negative Breast Cancer From the GeparQuinto Study. J Clin Oncol 2018;36:2281-7. [Crossref] [PubMed]
- Molina-Montes E, Pérez-Nevot B, Pollán M, et al. Cumulative risk of second primary contralateral breast cancer in BRCA1/BRCA2 mutation carriers with a first breast cancer: A systematic review and meta-analysis. Breast 2014;23:721-42. [Crossref] [PubMed]
- Rong G, Yi Z, Ma F, et al. DNA damage response as a prognostic indicator in metastatic breast cancer via mutational analysis. Ann Transl Med 2021;9:220. [Crossref] [PubMed]
- Xie N, Tian C, Wu H, et al. FGFR aberrations increase the risk of brain metastases and predict poor prognosis in metastatic breast cancer patients. Ther Adv Med Oncol 2020;12:1758835920915305. [Crossref] [PubMed]
- Zou Y, Zheng S, Xie X, et al. N6-methyladenosine regulated FGFR4 attenuates ferroptotic cell death in recalcitrant HER2-positive breast cancer. Nat Commun 2022;13:2672. [Crossref] [PubMed]
- Meric-Bernstam F, Bahleda R, Hierro C, et al. Futibatinib, an Irreversible FGFR1-4 Inhibitor, in Patients with Advanced Solid Tumors Harboring FGF/FGFR Aberrations: A Phase I Dose-Expansion Study. Cancer Discov 2022;12:402-15. [Crossref] [PubMed]
- Amina B, Lynda AK, Sonia S, et al. Fibroblast growth factor receptor 1 protein (FGFR1) as potential prognostic and predictive marker in patients with luminal B breast cancers overexpressing human epidermal receptor 2 protein (HER2). Indian J Pathol Microbiol 2021;64:254-60. [Crossref] [PubMed]
- An Y, Guan Y, Xu Y, et al. The diagnostic and prognostic usage of circulating tumor DNA in operable hepatocellular carcinoma. Am J Transl Res 2019;11:6462-74.
(English Language Editor: J. Jones)


