
Combined targeting of KRT23 and NCCRP1 as a potential novel therapeutic approach for the treatment of triple-negative breast cancer
Introduction
Breast cancer with triple-negative receptors lacks the genes for the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2)/neu. It accounts for 15–20% of all breast cancers and has a poor prognosis (1,2). In the absence of ER, PR, and HER2/neu, optimizing the therapeutic management of patients is difficult (2,3). Despite the improvements in combined surgical and radiochemotherapy (4-6), there remains a pressing need for new therapeutic approaches and molecular targets.
Keratin23 (KRT23) belongs to the acidic type I keratins; it is strongly expressed in colon adenocarcinomas but absent in normal colon mucosa. KRT23 knockdown decreases proliferation and affects the DNA damage response of colon cancer cells (7). Similarly, researchers have found that phosphoprotein KRT23 accumulates in microsatellite-stable (MSS) colon cancers in vivo and impacts the viability and proliferation in vitro (8). However, KRT23 has not been reported in triple-negative breast cancers (TNBC). Non-specific cytotoxic cell receptor 1 (NCCRP1) is a paralog of the F-box superfamily of proteins (ubiquitin ligases) that regulate the cell cycle (9,10). In particular, NCCRP1 can act as a prognostic signature in pancreatic cancer and squamous cell carcinoma (11,12). However, the role of NCCRP1 in TNBC is unknown.
In this study, we first assessed the roles of KRT23 and NCCRP1 in predicting the diagnosis and prognosis of TNBC using bioinformatics analysis based on the Clinical Proteomic Tumor Analysis Consortium (CPTAC). We then validated the messenger RNA (mRNA) and protein expressions of KRT23 and NCCRP1 in breast cancer. In addition to elucidating the molecular mechanisms regulating the biological actions of KRT23 and NCCRP1 in a breast cancer cell line, the purpose of the study was to investigate the effects of KRT23 and NCCRP1 on the proliferation of breast cancer cells. We present the following article in accordance with the MDAR reporting checklist (available at https://gs.amegroups.com/article/view/10.21037/gs-22-486/rc).
Methods
Data retrieval and determination of the prognostic value
We obtained data on gene expression and clinical characteristics using the Clinical Proteomic Tumor Analysis Consortium (CPTAC) (https://cptac-data-portal.georgetown.edu/cptac) and The Cancer Genome Atlas (TCGA) database (https://tcga-data.nci.nih.gov/tcga). Differential gene screening between TNBC and non-triple-negative breast cancers (NTNBC) was performed according to the “edgeR” filter criteria (log2|fold change| >0.5, false discovery rate (FDR) <0.05). We then analyzed the obtained differential genes (Gene Ontology) GO and (Kyoto Encyclopedia of Genes and Genomes) KEGG enrichment pathways.
This study used univariate and multivariate Cox regression analyses to construct a risk model and identify prognosis-related genes. Our survival analysis was based on the Kaplan-Meier analysis (KM). XCELL (13), TIMER (14), EPIC (15), QUANTISEQ (16), MCPCOUNTER (17), CIBERSORT-ABS (18), and CIBERSORT (18) software were used to determine the extent of tumor immune cell infiltration. To evaluate the clinical responses to breast cancer treatment, the half maximal inhibitory concentration (IC50s) of common chemotherapeutics were calculated using “pRRophetic” and “ggplot2”.
Clinical specimens and approval
Breast cancer tissues and corresponding adjacent normal tissues were collected from 29 patients who underwent surgical resection at the First Affiliated Hospital of Gannan Medical University, China. After surgical removal, the samples were immediately frozen in liquid nitrogen. This study was conducted according to the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of the First Affiliated Hospital of Gannan Medical University (No. LLSC-2022033101). Informed consent was obtained from the patients or their guardians.
Quantitative real-time polymerase chain reaction (qRT-PCR)
RNA was extracted using an RNA Extraction Kit (Qiagen, USA) according to the manufacturer’s instructions. Next, the RNA was reverse-transcribed using the TAKARA reverse transcription kit (TaKaRa, Dalian, China). TaKaRa SYBR Premix Ex TaqTM II Kit (TaKaRa, Dalian, China) was used to amplify the complementary DNA (cDNA)templates by qRT-PCR. The ABI 7500 Real-Time PCR System (Applied Biosystems, CA, USA) was used to perform qRT-PCR. The primer sequences are listed in Table S1.
Cell cultures and lentiviral transfection
Human breast cancer cells (MDA-MB-231 cell lines) were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The MDA-MB-231 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% Foetal Bovine Serum (FBS), 100 U/mL penicillin, and 100 mU/mL streptomycin (Corning, USA).
Overexpression of KRT23 and NCCRP1 (lv-KRT23 and lv-NCCRP1) were constructed and transfected in MDA-MB-231 cells using lentiviral overexpression plasmids Ubi-MCS-SV40-puromycin. Cells transfected with lv-negative control (NC) lentivirus served as a negative control. Next, we used the hU6-MCS-CMV-puromycin plasmid to knock down KRT23 and NCCRP1 in cells (sh-KRT23 and sh-NCCRP1) before transfection in MDA-MB-231 cells, with sh-NC cells used as a negative control. The culture was performed in 5% carbon dioxide (CO2) incubators (Thermos, USA). The interfering nucleotide sequence was designed according to the Invitrogen RNA interference sequence design website (https://rnaidesigner.thermofisher.com/). The RNA interference target sequences are presented in Table S2.
Immunohistochemistry and western blot (WB) assays
Immunohistochemistry was performed according to the manufacturer's instructions (Solarbio, Beijing, China). Sections were observed, and images were captured under a Leica DMRB microscope (Leica Microsystems, Germany). An extraction kit for total proteins (Keygen, Nanjing, China) was used to extract proteins from cells. Protein concentrations were determined using the BCA-100 Protein Quantitative Analysis Kit (Shanghai, China). WB was performed according to the standard protocols, and WB analysis was conducted using the antibodies listed in Table S3.
Cell Counting Kit 8 (CCK8) and 5-Ethynyl-2'-deoxyuridine (EdU) Cell Proliferation Kit
Briefly, MDA-MB-231 cells (1×103) were seeded into 96-well plates. At various time points (0, 1, 2, 3, and 4 days), we determined the value of absorbance (at 450 nm) for each cell according to the instruction manuals (Dojindo, Japan). To conduct the EdU assay, we followed the instructions provided with the EdU Cell Proliferation Assay Kit (RiboBio Co., Ltd., Guangzhou, Guangdong, China). Finally, the EdU-stained cells were examined under a confocal microscope (Carl Zeiss, Germany).
Dual-luciferase reporter assay
To explore the effect of SOX10 on the KRT23 and NCCRP1 promoter activity, the sequences of the KRT23 and NCCRP1 promoters were sub-cloned downstream of the luciferase reporter gene to create pGL3-KRT23 pro-Wild-type (Wt) and pGL3-NCCRP1 pro-Wild-type (Wt) plasmids. To test the binding specificity, a corresponding mutant was created with a changed region binding site to create pGL3-KRT23 pro-Mut-type (Mut) and pGL3-NCCRP1 pro-Mut-type (Mut) plasmids. The sequences of the wild-type KRT23 and NCCRP1 promoters were obtained from the UCSC genome browser (http://genome.ucsc.edu/). Luciferase activity was measured using a luciferase assay kit (K801–200, BioVision Inc., Milpitas, CA, USA). The relative luciferase activity was calculated based on firefly fluorescence versus Renilla fluorescence.
Statistical analysis
All statistical analyses were performed by using R software packages version 3.4.2. Experimental data analysis was performed using GraphPad Prism 8, with P<0.05 considered statistically significant.
Results
Identification of potential key genes associated with the prognosis of TNBC
We determined the differential expression of proteins between TNBC and NTNBC based on the CPTAC database (available at https://cdn.amegroups.cn/static/public/gs-22-486-1.xlsx). Next, we analyzed the differential proteins by GO and KEGG enrichment pathways. GO enrichment analysis revealed five significantly enriched GO terms (carboxylic acid biosynthetic process, gland development, gland morphogenesis, organic acid biosynthetic, and urogenital system development) (P<0.05) (Figure 1A). The KEGG enrichment analysis identified five significantly enriched KEGG pathways (arginine biosynthesis, estrogen signaling pathway, fatty acid metabolism, prostate cancer, and staphylococcus aureus infection) (P<0.05) (Figure 1B).
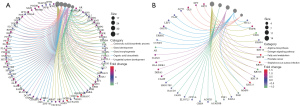
All differential proteins were analyzed to identify prognosis-related genes in triple-negative breast cancer. The univariate and multivariate Cox regression analyses found that KRT23 and NCCRP1 could independently predict prognosis (Figure 2A,2B), with high KRT23 and NCCRP1 expression levels indicating poor prognosis in triple-negative breast cancer (Figure 2C,2D). Immunohistochemistry showed that KRT23 and NCCRP1 had a high expression in TNBC tissues (Figure 2E).
Next, the risk score was calculated for each of the 105 patients based on the KRT23 and NCCRP1 expression levels from CPTAC, and the patients were then divided into high- and low-risk groups based on a cutoff value (the median risk score). An analysis of the Kaplan-Meier survival data revealed that the high-risk group had worse overall survival (OS), which suggests that the risks may have a prognostic value (Figure 2F).
High KRT23 and NCCRP1 protein levels promote breast cancer cell proliferation
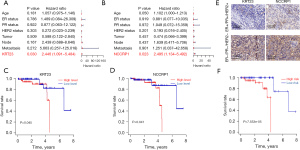
To further explore the biological role of KRT23 and NCCRP1 in breast cancer cells, the expressions of KRT23 and NCCRP1 were knocked down or overexpressed in MDA-MB231 cells using lentivirus (Figure 3A,3B). The EdU and CCK-8 assays showed that silenced KRT23 or NCCRP1 restored cell proliferation (Figure 3C-3E). However, the overexpression of KRT23 or NCCRP1 promoted cell proliferation (Figure 3F-3H). These results suggested that KRT23 or NCCRP1 might be essential oncogenes in TNBC.
SOX10 regulates KRT23 and NCCRP1 gene expression in MDA-MB231 cells
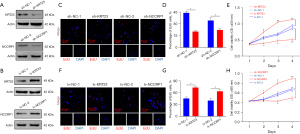
The Venn diagram analysis identified the differentially expressed transcription factors, which co-regulated KRT23 or NCCRP1 expression in TNBC. The results showed that KRT23 and NCCRP1 had 12 differentially expressed common transcription factors (Figure 4A). An overall correlation between the 14 features is displayed in a heatmap, and a significant correlation was found between SOX10 expression and KRT23 or NCCRP1 expression (Figure 4B).
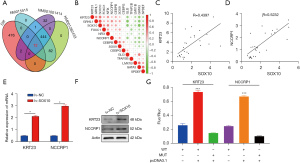
Additionally, we found that SOX10 was positively correlated with KRT23 and NCCRP1 in TNBC tissues (Figure 4C,4D). Meanwhile, KRT23 and NCCRP1 expression was promoted by SOX10 overexpression (Figure 4E,4F). Dual-luciferase reporter assays in MDA-MB231 cells were performed to validate the regulation of KRT23 and NCCRP1 expression by SOX10 (Figure 4G).
The risk score was associated with immune cell infiltration and chemotherapeutics
In this study, multiple software programs (XCEL, TIMER, QUANTISEQ, MCPCOUNTER, EPIC, CIBERSORT-ABS, CIBERSORT) were used to analyze the relationships between patients in the high- and low-risk groups and immune cells in the risk prediction model (Figure 5A). To determine whether the chemotherapeutic treatment efficacy was correlated with the risk scores, we examined the relationship between risk scores and IC50.
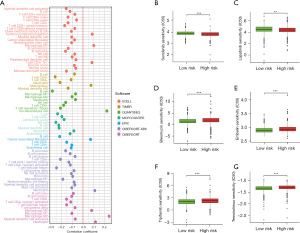
In the present study, we sought to determine the association between the risk score and the efficacy of common chemotherapeutic agents in the treatment of breast cancer in TCGA database. Our results revealed that a high-risk score was related to a lower half maximal inhibitory concentration (IC50) of chemotherapeutics such as Sorafenib (Figure 5B) and Lapatinib (Figure 5C), while a low-risk score was related to a lower IC50 of chemotherapeutics such as Bleomycin (Figure 5D), Embelin (Figure 5E), Tipifarnib (Figure 5F), and Temsirolimus (Figure 5G). These results indicated that the model was a potential predictor of chemosensitivity.
Discussion
Breast cancers characterized by triple-negative status tend to be more malignant and have a poorer prognosis (1,2,4-6,19,20). To decrease the mortality rate and improve the prognosis of breast cancer patients, a model for predicting breast cancer risk should be developed. In this study, a risk model was constructed and validated using the CPTAC databases.
Using multivariate and univariate Cox regression analyses, an assessment model was established for KRT23 and NCCRP1. We found that high expression levels of KRT23 and NCCRP1 were indicative of poor prognosis and that the gene expression patterns of multiple genes are considerably more predictive of future risks and have a higher level of consistency when assessing risk. KRT23 is a newly discovered member of the keratin family. Various tumor tissues, including pancreatic cancer (21), colorectal carcinoma (7,22-24), and hepatocellular carcinoma (25,26), have exhibited aberrant expression of KRT23. Our findings support the same results, namely that the expression of KRT23 was increased in TNBC and promoted cell proliferation. In this study, we investigated the expression and functions of NCCRP1 in cells. Our findings also found the same; the expression of NCCRP1 was increased in TNBC and promoted cell proliferation. Therefore, we can design specific antibodies against KRT23 and NCCRP1 to block their effects on breast cancer proliferation.
To identify the transcription factors regulating the joint regulation of KRT23 and NCCRP1, we found their transcripts through the UCSC database and performed transcription factor prediction on all transcripts. Venn diagram analysis identified 12 differentially expressed transcription factors, which co-regulated KRT23 or NCCRP1 expression in TNBC. We confirmed our hypothesis that SOX10 regulated KRT23 and NCCRP1.
We also analyzed the risk scores associated with immune cell infiltration and chemotherapeutics. The XCEL database showed a strong association with the common lymphoid progenitor, M2 macrophages, and CD4+ Th1 T cells in high-risk group patients. The TIMER database showed a strong association with macrophages in high-risk group patients. The QUANTISEQ database demonstrated a strong association with uncharacterized cells in high-risk group patients. The EPIC database showed a strong association with cancer-associated fibroblasts in high-risk group patients. The CIBERSORT-ABS database showed a strong association with neutrophils in high-risk group patients. The CIBERSORT database showed a strong association with M2 macrophages and neutrophils in high-risk group patients. Based on these results, we found that high-risk patients were closely associated with M2 type macrophage infiltration. Therefore, in the treatment of triple negative breast cancer, it can be combined with M2 macrophage inhibitor to improve the efficacy of chemotherapy. Similarly, we can also predict the relationship between KRT23 and NCCRP1 and immune cells by analyzing the correlation between KRT23 and NCCRP1 and immune-related genes.
The sensitivity to chemotherapy was different between the high- and low-risk groups. In conclusion, these results indicated that the model was a potential predictor of chemosensitivity.
Acknowledgments
We thank all of the participants in the present research. We also thank the reviewers for their valuable advice.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://gs.amegroups.com/article/view/10.21037/gs-22-486/rc
Data Sharing Statement: Available at https://gs.amegroups.com/article/view/10.21037/gs-22-486/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://gs.amegroups.com/article/view/10.21037/gs-22-486/coif). The authors have no conflicts of interest to declare
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted according to the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of the First Affiliated Hospital of Gannan Medical University (No. LLSC-2022033101). Informed consent was obtained from the patients or their guardians.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Li K, Zong D, Sun J, et al. Rewiring of the Endocrine Network in Triple-Negative Breast Cancer. Front Oncol 2022;12:830894. [Crossref] [PubMed]
- Waks AG, Winer EP. Breast Cancer Treatment: A Review. JAMA 2019;321:288-300. [Crossref] [PubMed]
- Denkert C, Liedtke C, Tutt A, et al. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet 2017;389:2430-42. [Crossref] [PubMed]
- Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med 2010;363:1938-48. [Crossref] [PubMed]
- Xu X, Xie Q, Xie M, et al. LncRNA SNHG8 Serves as an Oncogene in Breast Cancer Through miR-634/ZBTB20 Axis. Cancer Manag Res 2021;13:3017-28. [Crossref] [PubMed]
- Wen X, Chen Y, Fang X. Overexpression of HOXD8 inhibits the proliferation, migration and invasion of breast cancer cells by downregulating ILP2 expression. Exp Ther Med 2021;22:1006. [Crossref] [PubMed]
- Birkenkamp-Demtröder K, Hahn SA, Mansilla F, et al. Keratin23 (KRT23) knockdown decreases proliferation and affects the DNA damage response of colon cancer cells. PLoS One 2013;8:e73593. [Crossref] [PubMed]
- Birkenkamp-Demtroder K, Mansilla F, Sørensen FB, et al. Phosphoprotein Keratin 23 accumulates in MSS but not MSI colon cancers in vivo and impacts viability and proliferation in vitro. Mol Oncol 2007;1:181-95. [Crossref] [PubMed]
- Cordero H, Guzmán-Villanueva LT, Chaves-Pozo E, et al. Comparative ontogenetic development of two marine teleosts, gilthead seabream and European sea bass: New insights into nutrition and immunity. Dev Comp Immunol 2016;65:1-7. [Crossref] [PubMed]
- Kallio H, Tolvanen M, Jänis J, et al. Characterization of non-specific cytotoxic cell receptor protein 1: a new member of the lectin-type subfamily of F-box proteins. PLoS One 2011;6:e27152. [Crossref] [PubMed]
- Zuo H, Chen L, Li N, et al. Identification of a Ubiquitination-Related Gene Risk Model for Predicting Survival in Patients With Pancreatic Cancer. Front Genet 2020;11:612196. [Crossref] [PubMed]
- Miwa T, Kanda M, Koike M, et al. Identification of NCCRP1 as an epigenetically regulated tumor suppressor and biomarker for malignant phenotypes of squamous cell carcinoma of the esophagus. Oncol Lett 2017;14:4822-8. [Crossref] [PubMed]
- Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. [Crossref] [PubMed]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509-14. [Crossref] [PubMed]
- Racle J, Gfeller D. EPIC: A Tool to Estimate the Proportions of Different Cell Types from Bulk Gene Expression Data. Methods Mol Biol 2020;2120:233-48. [Crossref] [PubMed]
- Plattner C, Finotello F, Rieder D. Deconvoluting tumor-infiltrating immune cells from RNA-seq data using quanTIseq. Methods Enzymol 2020;636:261-85. [Crossref] [PubMed]
- Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol 2016;17:218. [Crossref] [PubMed]
- Chen B, Khodadoust MS, Liu CL, et al. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol 2018;1711:243-59. [Crossref] [PubMed]
- Liao R, Chen X, Cao Q, et al. HIST1H1B Promotes Basal-Like Breast Cancer Progression by Modulating CSF2 Expression. Front Oncol 2021;11:780094. [Crossref] [PubMed]
- Xie R, Wang M, Zhou W, et al. Long Non-Coding RNA (LncRNA) UFC1/miR-34a Contributes to Proliferation and Migration in Breast Cancer. Med Sci Monit 2019;25:7149-57. [Crossref] [PubMed]
- Zhang JS, Wang L, Huang H, et al. Keratin 23 (K23), a novel acidic keratin, is highly induced by histone deacetylase inhibitors during differentiation of pancreatic cancer cells. Genes Chromosomes Cancer 2001;30:123-35. [Crossref] [PubMed]
- Zhang N, Zhang R, Zou K, et al. Keratin 23 promotes telomerase reverse transcriptase expression and human colorectal cancer growth. Cell Death Dis 2017;8:e2961. [Crossref] [PubMed]
- Gao X, Yang J. Identification of Genes Related to Clinicopathological Characteristics and Prognosis of Patients with Colorectal Cancer. DNA Cell Biol 2020;39:690-9. [Crossref] [PubMed]
- Qin Y, Chen L, Chen L. Identification and verification of key cancer genes associated with prognosis of colorectal cancer based on bioinformatics analysis. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2021;46:1063-70. [PubMed]
- Wang K, Xu X, Nie Y, et al. Identification of tumor-associated antigens by using SEREX in hepatocellular carcinoma. Cancer Lett 2009;281:144-50. [Crossref] [PubMed]
- Kim D, Brocker CN, Takahashi S, et al. Keratin 23 Is a Peroxisome Proliferator-Activated Receptor Alpha-Dependent, MYC-Amplified Oncogene That Promotes Hepatocyte Proliferation. Hepatology 2019;70:154-67. [Crossref] [PubMed]


